A performance assessment of relatedness inference methods using genome-wide data from thousands of relatives
|
|
|
- Elfrieda Mills
- 5 years ago
- Views:
Transcription
1 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not A performance assessment of relatedness inference methods using genome-wide data from thousands of relatives Monica D. Ramstetter,, Thomas D. Dyer, Donna M. Lehman, Joanne E. Curran, Ravindranath Duggirala, John Blangero, Jason G. Mezey,3, and Amy L. Williams, Department of Biological Statistics and Computational Biology, Cornell University, Ithaca, NY 4853, USA South Texas Diabetes and Obesity Institute, University of Texas Rio Grande Valley, Brownsville, TX 7850, USA and Edinburg, TX 78539, USA 3 Department of Genetic Medicine, Weill Cornell Medicine, New York, NY 0065, USA Abstract Inferring relatedness from genomic data is an essential component of genetic association studies, population genetics, forensics, and genealogy. While numerous methods exist for inferring relatedness, thorough evaluation of these methods in real data has been lacking. Here, we report an assessment of state-ofthe-art relatedness inference methods using a dataset with,485 individuals contained in several large pedigrees that span up to six generations. We find that all methods have high accuracy ( 93% 99%) when reporting first and second degree relationships, but their accuracy dwindles to less than 60% for fifth degree relationships. However, the inferred relationships were correct to within one relatedness degree at a rate of 83% 99% across all methods and considered relationship degrees. Furthermore, most methods infer unrelated individuals correctly at a rate of 99%, suggesting a low rate of false positives. Overall, the most accurate methods were ERSA.0 and approaches that classify relationships using the IBD segments inferred by Refined IBD and IBDseq. Combining results from the most accurate methods provides little accuracy improvement, indicating that novel approaches for relatedness inference may be needed to achieve a sizeable jump in performance. The recent explosive growth in sample sizes of genetic datasets has led to an increasing proportion of close relatives hidden within these large studies, necessitating relatedness detection. Inferring relatedness between samples 3 is an essential step in performing genetic association studies 4 6 and linkage analysis 7 9, is a powerful tool for forensic genetics,0,, and is needed to account for or remove relatives in population genetic analyses 4. Relatedness estimation has also drawn the interest of the general public via companies such as 3andMe and AncestryDNA which advertise their ability to find and report relatives, allowing individuals to explore their ancestry and genealogy. The broad utility of relatedness estimation has motivated the development of numerous methods for such inference. These methods work by estimating the proportion of the genome shared identical by descent (IBD) between individuals,3 or a closely-related quantity, where an allele in two or more individuals genomes is said to be IBD if those individuals inherit it from a recent common ancestor. As previously shown, the distributions of IBD for different relatedness classes (such as first cousins and half-first cousins) are expected to overlap,5, posing a challenge for these inference procedures. Here, we present a rigorous evaluation of state-of-the-art methods that can scale to large study sizes, including seven that directly infer genome-wide relatedness measures 6 and four IBD segment detection methods 3 6 that we utilized to infer these quantities. To assess each of these methods, we used SNP array genotypes from Mexican American individuals contained in large pedigrees from the San Antonio Mexican American Family Studies (SAMAFS) 7 9. Our analysis sample included,485 individuals genotyped at 5,84 SNPs (Supplemental Note) within pedigrees that span up to six generations with genotype data Correspondence: mdr3@cornell.edu (M.D.R.), alw89@cornell.edu (A.L.W)
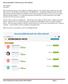
2 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not Degree Number of Pairs 4,969 6, ,44 4 7, ,50 Unrelated 3,05,035 Total 3,057,89 Table : Numbers of pairs of individuals from the SAMAFS dataset reported to have relatedness between first and fifth degree and counts of unrelated pairs used for the evaluation. Only individuals from distinct pedigrees are considered unrelated. from as many as five generations of individuals. Given this large sample, including 3 pedigrees with >50 individuals (Supplemental Figure ), numerous close relatives exist, and we used these to evaluate each of the inference methods. In particular, there are >4,500 pairs of individuals within each of the first through fifth degree relatedness classes that we evaluated, and we further considered more than three million pairs of individuals that are in distinct pedigrees and hence assumed unrelated (Table ). Prior analyses of relatedness inference methods considered either simulated data 7,8,0 which may not fully capture the complexities of real data or used small sample sizes 7,8,,30. Our analysis using real data for large numbers of up to fifth degree relatives provides a comprehensive evaluation of these relatedness inference methods. Our analysis considered each method s ability to correctly infer the degree of relatedness between the pairs of samples based on their reported relationships. These reported relationships are extremely reliable and in most cases we can validate them via first degree connections among samples in the densely-genotyped SAMAFS pedigrees. Some methods directly infer the degree of relatedness 9 while others infer a kinship coefficient 7,8,0, a coefficient of relatedness 6, (which is two times the kinship coefficient 3 ), or instead detect IBD segments 3 6 (Table ). To infer the degree of relatedness from an estimated kinship coefficient for a pair of samples, we use the ranges of estimated kinship values from the KING method 7 (Table 3). These ranges use differences in powers of two for the relatedness degree intervals, which is generally consistent with simulations 3. For IBD detection methods that report the number of IBD segments shared at a locus 3,6 denoted IBD0, IBD, and IBD for the corresponding number of copies that are IBD it is straightforward to calculate a kinship coefficient. This coefficient, φ ij, between a pair of samples i, j denotes the probability that a randomly selected allele in individual i is IBD with a randomly selected allele from the same genomic position in j. Let p () ij and p () ij denote the proportion of their genomes that individuals i, j share IBD and IBD respectively; then the kinship coefficient is φ ij = p() ij 4 + p() ij. The p() ij and p () ij are simply the sum of the genetic lengths of the IBD and IBD segments, respectively, between samples i, j divided by the total genetic length of the genome analyzed. (Note if i = j, then φ ii = ( + f i) where f i is the kinship coefficient between the parents of i which is equivalent to the inbreeding coefficient of individual i.) For the IBD detection methods that do not distinguish between regions that are IBD from IBD 4,5, the proportion of the genome that is inferred to be IBD0 provides an alternate means of estimating the degree of relatedness (Table 3), with the ranges of values here again from the KING paper 7. We classified individuals with lower kinship coefficients or higher IBD0 rates than indicated for the fifth degree range as unrelated. Using the SAMAFS sample, we assessed the performance of each program by using them to classify all pairs of individuals. Figure shows the proportion of sample pairs inferred to be within each of the degree classes that we considered (first through fifth degree and unrelated), with results separated according to the reported and inferred relatedness degrees of the pairs. All methods perform well when inferring first and second degree relatives, with the accuracy ranging from 98.4% to 99.5% for first degree relatives, and from 93% to 98.6% for second degree relatives. For more distant relatedness, the IBD-based methods have
3 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not 3 Method Version Citation Number Type Output Parallelized? Runtime ( cores used if >) Requires independent markers Input required from outside program Accounts for population structure ERSA.0 9 IBD segment-based Degree of relatedness N 4.5h N IBD segments NA fastibd Beagle IBD segment-finding IBD segments N 55.5h N NA NA GERMLINE (-haploid).5. 3 IBD segment-finding (Distinguishes IBD and IBD) IBD segments N 0m N Phased genotypes NA IBDseq r06 5 IBD segment-finding IBD segments Y 33.5h ( 6) N NA NA KING (KING-robust).4 7 IBD 0,, N 5m Y NA Y PC-Relate.0. IBD 0,, N 9h Y Pairwise kinship coefficients Y PLINK.9.90bk 6 IBD 0,, N 0s Y NA N PREST-plus 4. IBD 0,, N 79h N NA N REAP. 8 IBD 0,, N 4h Y Ancestral population Y Refined IBD Beagle 4. 6 IBD segment-finding (Distinguishes IBD and IBD) IBD segments Y 9h ( 6) N NA NA RelateAdmix 0. 0 IBD 0,, Y 6h ( 6) Y Ancestral population Y Table : Properties of the relationship inference methods we analyzed. Type indicates the inference methodology the program uses. Runtime is wall clock time to run the program; we ran parallelized programs using the numbers of cores indicated in parentheses: total compute time for the parallelized programs is the runtime multiplied by the number of cores used. Input required from outside program indicates extraneous information needed to run the program. Programs that use either principal components or ancestral population are indicated as accounting for population structure. Y indicates yes, N indicates no, and NA indicates not applicable. Runtimes are from a machine with four AMD Opteron GHz processors (64 cores total) and 56 GB memory.
4 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not 4 Expected Accepted range for: Relationship Degree # Meiosis IBD0 IBD IBD φ φ P(IBD=0) Parent-child 0 0 (, ] 3/ / < 0. Full siblings (not MZ twin) ( ) /4 / /4 (, ] 3/ / [0., 0.365) Grandparent / / 0 (, ] [0.365, ) 3 5/ 3/ 3/ Avuncular 3 / / 0 (, ] [0.365, ) 3 5/ 3/ 3/ Double-cousins 4 ( ) 9/6 3/8 /6 (, ] [0.365, ) 3 5/ 3/ 3/ Half sibling / / 0 (, ] [0.365, ) 3 5/ 3/ 3/ First Cousin 3 4 3/4 /4 0 (, ] [, ) 4 7/ 5/ 3/ 5/ Double half-cousins 3 5 ( ) 3/3 7/3 /64 (, ] [, ) 4 7/ 5/ 3/ 5/ Great-grandparent 3 3 3/4 /4 0 (, ] [, ) 4 7/ 5/ 3/ 5/ Half-/grand-avuncular 3 4 3/4 /4 0 (, ] [, ) 4 7/ 5/ 3/ 5/ First cousin once removed 4 5 7/8 /8 0 (, ] [, ) 5 9/ 7/ 5/ 7/ Great-great-grandparent 4 4 7/8 /8 0 (, ] [, ) 5 9/ 7/ 5/ 7/ Half-grand-/great-grandavuncular 4 5 7/8 /8 0 5 ( 9/, 7/ ] [ 5/, 7/ ) First cousin twice removed 5 6 5/6 /6 0 (, ] [, ) 6 / 9/ 7/ 9/ Second cousin 5 6 5/6 /6 0 (, ] [, ) 6 / 9/ 7/ 9/ GGG-grandparent 5 5 5/6 /6 0 (, ] [, ) 6 / 9/ 7/ 9/ Table 3: For a range of relationship types, the corresponding degree of relatedness of the individuals; the number of meioses that separate them, with ( ) indicating samples that are related along two lines of descent (such as full siblings) that have the listed meiotic distance on both lines; of the genome that are expected to be IBD0, IBD, and IBD between the samples; and expected kinship coefficient φ. For inferring a degree of relatedness from a kinship coefficient, the range of values that map to the given degree are listed. Likewise for inference using IBD0, the of IBD0 values that map to each degree are shown. The list does not include all possible relationship types for the degrees of relatedness listed. higher accuracy than those that rely on allele frequencies of independent markers for example, for fifth degree relatives, the top performing IBD-based method has 59.4% accuracy while the highest performing allele frequency-based method has only 53.8% accuracy. Overall, the most accurate programs are ERSA.0, Refined IBD, and IBDseq. The improved accuracy of IBD-based methods may be due to their focus on identifying long stretches of identical segments that more readily discriminate recent shared relatedness from chance sharing of alleles. Noting that the SAMAFS consist of admixed Mexican American individuals, we examined the accuracy results among the allele frequency-based methods, of which several account for population structure. Of all these methods, PC-Relate has the highest accuracy across all levels of relatedness, and it does account for population structure using principal components. Overall, the results are mixed with regards to accounting for population structure and accuracy, with PC-Relate, REAP, RelateAdmix, and KING all incorporating population structure into their models, and PREST-plus and PLINK ignoring this structure. Because relatedness structure can confound methods that detect population structure, we employed a procedure designed to locate true ancestral population for the input supplied to REAP and RelateAdmix (Supplemental Note). PC-Relate, by contrast, addresses these concerns by performing population structure analysis internally using a set of samples with low levels of relatedness. However, IBD detection methods do not directly account for population structure and generally have the best performance. The inference accuracy of all methods decreases for higher relatedness degrees, likely due to the exponential drop in mean pairwise IBD shared and an increased coefficient of variation as relatedness decreases 5,3,33. In particular, for fifth degree relatives, the accuracy rates for all methods are very low at less than 60%. However, in nearly all cases ( 83.8%), the programs correctly inferred the degree of relatedness to within
5 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not 5 one degree of that reported in the SAMAFS pedigrees. IBDseq has the highest within-one-degree accuracy for reported fourth degree pairs (the relationship class with the lowest accuracies for off-by-one inference) at 98.7%. At the same time, the methods classify an average of 97.9% of pairs of unrelated individuals correctly, averaged across all programs (99.7% when PLINK is excluded), with few instances of fifth or greater degree of relatedness inferred for these pairs. These results suggest that, when methods do detect relatedness even as far distant as fifth degree the individuals are likely to be truly related. Because the SAMAFS data consist of many closely related individuals, the allele frequencies derived from it have the potential to be biased. Furthermore, haplotype phasing and therefore IBD inference accuracy might be greater than would be achieved in a more outbred sample. To ensure the performance results presented here also apply to analyses of non-pedigree datasets, we identified a set of unrelated individuals using FastIndep 34 and merged these samples with pairs of related individuals to form,000 datasets that include different pairs of relatives (Supplemental Note). Each reduced dataset contains at most one pair of samples from any distinct SAMAFS pedigree, limiting the potential for bias. When classifying the related individuals included in at least one of these reduced datasets, PLINK s inference accuracy differs by less than 3% compared to the full dataset (Supplemental Figure ), suggesting that allele frequency biases are small and only minimally impact inference accuracy. In order to test the IBD detection methods, we further merged 580 HapMap samples 35 with each of the reduced datasets (Supplemental Note). Results from running IBD detection methods on these datasets show a reduction in accuracy that ranges between 0% 8%, yet the results are still consistent with those of the larger analysis (Supplemental Figure 3). Specifically, the IBD segment-finding methods tend to have higher performance than allele frequency-based methods, supporting the conclusion that IBD segment-based methods provide the highest accuracy. This is true even in the reduced datasets that have no more than,04 samples and therefore are subject to a relatively high level of phasing errors. We examined the pairs of samples that were inferred to be related but were reported as unrelated (in distinct pedigrees) in the SAMAFS dataset. ERSA.0, Refined IBD, and IBDseq all inferred a small number of first through third degree relationships that connect individuals from different pedigrees within SAMAFS (Figure ). Overall, we found 48 pairs of pedigrees with at least five pairs of relatives between them which all three methods unanimously infer to have the same degree of relatedness. Additionally, these three methods agreed on the inference of 374 and,63 pairs of fourth and fifth degree relatives between the pedigrees (not shown). These results highlight the importance of checking for relatedness among samples in all cohorts, and indicate that there can be sizable numbers of relatives across a range of degrees even in well-studied samples. As current methods provide only moderate accuracy when classifying third through fifth degree relatives, we evaluated the potential for increasing performance by combining inference results from the top three programs. We used an approach that calls the degree of relatedness for a pair only when all three programs unanimously agree on the relatedness degree, providing no classification for other pairs. The resulting inference accuracy increased only negligibly (0.5%, 0.%,.6%, 3.%,.8%, and 0.0%, respectively for first through fifth degree and unrelated pairs) in comparison to the most accurate method s performance in each degree class. We also considered a majority vote between the three programs, discarding the cases in which all three programs inferred a different degree (only two cases were of this class). With this approach, there is a slight decrease in performance overall (-0.46%, -0.6%, -.4%, -.5%, +0.8%, +0.0%). These results suggest that while there is room for improvement in the specificity of relatedness inference methods, dramatic improvement is likely to be achieved only with novel approaches and not composites of current methods. We have presented a detailed comparison of state-of-the-art relatedness inference methods using thousands of pairs of individuals that range from first to fifth degree relatives as well as numerous individuals that are reported to be unrelated. All the methods we assessed reliably identify first and second degree relatives as well as unrelated pairs (accuracy 93% 99%), but their accuracy falls precipitously when classifying third to fifth degree relatives. This is unsurprising given the increased coefficient of variation as well as greater skewness in the proportion of genome shared as the meiotic distance between two relatives increases. Despite these challenges, the inferred relationship was within one degree of the reported relationship at a rate of 83% 99% for all programs and relationship degrees (Figure ). Misreported or unknown relationships in
6 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not 6 the SAMAFS dataset likely explain some of the inference errors, particularly since even some confidently inferred first degree relationships were likely misreported as a more distant relationship (Supplemental Table 4) or as unrelated (Figure ). We find that IBD-based methods outperform other approaches for more distantly-related pairs, though notably these packages require substantially more compute time to run which may limit their utility in some applications (Table ). While the precise performance results presented here are specific to the SAMAFS sample, we find that reducing the sample size still produces similar results, with methods that leverage IBD segments having greater accuracy than other approaches. Therefore, the results presented here should be generalizable and indicate overall properties of relationship inference methodologies: approaches that use IBD segments outperform other methods for third degree and more distant relatives; and the specificity of relatedness inference, even in a dataset where phase accuracy may be relatively high, is inhibited for all but the closest relatives.
7 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not 7 Figure : Performance comparison of the evaluated methods using the SAMAFS dataset. Bar plots indicate the percentage of pairs of samples that are reported to have a given degree of relatedness and who are inferred to be in each degree class. The bar plots are separated on the horizontal axis by the reported relatedness degree and on the vertical axis by inferred relatedness degree. For clarity, the plots list above each bar the percentage number that the corresponding bar depicts. Program names listed in red are IBD-based methods while those in black utilize allele frequencies for inference.
8 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not 8 Figure : Relationships discovered between individuals from different SAMAFS pedigrees. Bands on the perimeter of the elliptical plot indicate distinct pedigrees within SAMAFS with band size proportional to the number of individuals in the pedigree. Curves between two bands correspond to discovered relative pairs with color indicating the degree of relatedness: red for first degree, green for second degree, and blue for third degree. Points where the curves end correspond to specific individuals, and a single point may have multiple curves running to it, indicating several relationships between that individual and others in the dataset.
9 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not REFERENCES 9 References [] Bruce S Weir, Amy D Anderson, and Amanda B Hepler. Genetic relatedness analysis: modern data and new challenges. Nature Reviews Genetics, 7(0):77 780, 006. [] Elizabeth A Thompson. Identity by descent: variation in meiosis, across genomes, and in populations. Genetics, 94():30 36, 03. [3] Doug Speed and David J Balding. Relatedness in the post-genomic era: is it still useful? Nature Reviews Genetics, 6():33 44, 05. [4] Jonathan Marchini, Lon R Cardon, Michael S Phillips, and Peter Donnelly. The effects of human population structure on large genetic association studies. Nature Genetics, 36(5):5 57, 004. [5] Joel N Hirschhorn and Mark J Daly. Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics, 6():95 08, 005. [6] Benjamin F Voight and Jonathan K Pritchard. Confounding from cryptic relatedness in case-control association studies. PLOS Genetics, (3):e3, 005. [7] Jeffrey R O Connell and Daniel E Weeks. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. American Journal of Human Genetics, 63():59 66, 998. [8] Jurg Ott. Analysis of human genetic linkage. JHU Press, 999. [9] Michael P Epstein, William L Duren, and Michael Boehnke. Improved inference of relationship for pairs of individuals. American Journal of Human Genetics, 67(5):9 3, 000. [0] Mark A Jobling and Peter Gill. Encoded evidence: DNA in forensic analysis. Nature Reviews Genetics, 5(0):739 75, 004. [] Manfred Kayser and Peter de Knijff. Improving human forensics through advances in genetics, genomics and molecular biology. Nature Reviews Genetics, (3):79 9, 0. [] David C Queller and Keith F Goodnight. Estimating relatedness using genetic markers. Evolution, pages 58 75, 989. [3] Laurence D Hurst. Genetics and the understanding of selection. Nature Reviews Genetics, 0():83 93, 009. [4] Joshua G Schraiber and Joshua M Akey. Methods and models for unravelling human evolutionary history. Nature Reviews Genetics, 6():77 740, 05. [5] WG Hill and BS Weir. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genetics Research, 93(0):47 64, 0. [6] Christopher C Chang, Carson C Chow, Laurent CAM Tellier, Shashaank Vattikuti, Shaun M Purcell, and James J Lee. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience, 4():, 05. [7] Ani Manichaikul, Josyf C Mychaleckyj, Stephen S Rich, Kathy Daly, Michèle Sale, and Wei-Min Chen. Robust relationship inference in genome-wide association studies. Bioinformatics, 6(): , 00. [8] Timothy Thornton, Hua Tang, Thomas J Hoffmann, Heather M Ochs-Balcom, Bette J Caan, and Neil Risch. Estimating kinship in admixed populations. American Journal of Human Genetics, 9(): 38, 0. [9] Hong Li, Gustavo Glusman, Hao Hu, et al. Relationship estimation from whole-genome sequence data. PLOS Genetics, 0(), 04.
10 biorxiv preprint first posted online Feb. 4, 07; doi: The copyright holder for this preprint (which was not REFERENCES 0 [0] Ida Moltke and Anders Albrechtsen. RelateAdmix: a software tool for estimating relatedness between admixed individuals. Bioinformatics, 30(7):07 08, 04. [] Lei Sun and Apostolos Dimitromanolakis. PREST-plus identifies pedigree errors and cryptic relatedness in the GAW8 sample using genome-wide SNP data. BMC Proceedings, 8(Suppl ):S3, 04. [] Matthew P Conomos, Alexander P Reiner, Bruce S Weir, and Timothy A Thornton. Model-free estimation of recent genetic relatedness. American Journal of Human Genetics, 98():7 48, 06. [3] Alexander Gusev, Jennifer K Lowe, Markus Stoffel, Mark J Daly, David Altshuler, Jan L Breslow, Jeffrey M Friedman, and Itsik Pe er. Whole population, genome-wide mapping of hidden relatedness. Genome Research, 9():38 36, 009. [4] Brian L Browning and Sharon R Browning. A fast, powerful method for detecting identity by descent. American Journal of Human Genetics, 88():73 8, 0. [5] Brian L Browning and Sharon R Browning. Detecting identity by descent and estimating genotype error rates in sequence data. American Journal of Human Genetics, 93(5):840 85, 03. [6] Brian L Browning and Sharon R Browning. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics, 94():459 47, 03. [7] Braxton D Mitchell, Candace M Kammerer, John Blangero, Michael C Mahaney, David L Rainwater, Bennett Dyke, James E Hixson, Richard D Henkel, R Mark Sharp, Anthony G Comuzzie, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. Circulation, 94(9):59 70, 996. [8] Ravindranath Duggirala, John Blangero, Laura Almasy, Thomas D Dyer, Kenneth L Williams, Robin J Leach, Peter O Connell, and Michael P Stern. Linkage of type diabetes mellitus and of age at onset to a genetic location on chromosome 0q in Mexican Americans. American Journal of Human Genetics, 64(4):7 40, 999. [9] Kelly J Hunt, Donna M Lehman, Rector Arya, Sharon Fowler, Robin J Leach, Harald HH Göring, Laura Almasy, John Blangero, Tom D Dyer, Ravindranath Duggirala, et al. Genome-wide linkage analyses of type diabetes in Mexican Americans. Diabetes, 54(9):655 66, 005. [30] Chad D Huff, David J Witherspoon, Tatum S Simonson, Jinchuan Xing, W Scott Watkins, Yuhua Zhang, Therese M Tuohy, Deborah W Neklason, Randall W Burt, Stephen L Guthery, et al. Maximumlikelihood estimation of recent shared ancestry (ERSA). Genome Research, (5): , 0. [3] Sewall Wright. Coefficients of inbreeding and relationship. The American Naturalist, 56(645): , 9. [3] William G Hill. Variation in genetic identity within kinships. Heredity, 7:65 653, 993. [33] Peter M Visscher. Whole genome approaches to quantitative genetics. Genetica, 36():35 358, 009. [34] Kuruvilla Joseph Abraham and Clara Diaz. Identifying large sets of unrelated individuals and unrelated markers. Source code for biology and medicine, 9():, 04. [35] International HapMap 3 Consortium et al. Integrating common and rare genetic variation in diverse human populations. Nature, 467(73):5 58, 00.
University of Washington, TOPMed DCC July 2018
 Module 12: Comput l Pipeline for WGS Relatedness Inference from Genetic Data Timothy Thornton (tathornt@uw.edu) & Stephanie Gogarten (sdmorris@uw.edu) University of Washington, TOPMed DCC July 2018 1 /
Module 12: Comput l Pipeline for WGS Relatedness Inference from Genetic Data Timothy Thornton (tathornt@uw.edu) & Stephanie Gogarten (sdmorris@uw.edu) University of Washington, TOPMed DCC July 2018 1 /
ville, VA Associate Editor: XXXXXXX Received on XXXXX; revised on XXXXX; accepted on XXXXX
 Robust Relationship Inference in Genome Wide Association Studies Ani Manichaikul 1,2, Josyf Mychaleckyj 1, Stephen S. Rich 1, Kathy Daly 3, Michele Sale 1,4,5 and Wei- Min Chen 1,2,* 1 Center for Public
Robust Relationship Inference in Genome Wide Association Studies Ani Manichaikul 1,2, Josyf Mychaleckyj 1, Stephen S. Rich 1, Kathy Daly 3, Michele Sale 1,4,5 and Wei- Min Chen 1,2,* 1 Center for Public
Detection of Misspecified Relationships in Inbred and Outbred Pedigrees
 Detection of Misspecified Relationships in Inbred and Outbred Pedigrees Lei Sun 1, Mark Abney 1,2, Mary Sara McPeek 1,2 1 Department of Statistics, 2 Department of Human Genetics, University of Chicago,
Detection of Misspecified Relationships in Inbred and Outbred Pedigrees Lei Sun 1, Mark Abney 1,2, Mary Sara McPeek 1,2 1 Department of Statistics, 2 Department of Human Genetics, University of Chicago,
Kinship/relatedness. David Balding Professor of Statistical Genetics University of Melbourne, and University College London.
 Kinship/relatedness David Balding Professor of Statistical Genetics University of Melbourne, and University College London 2 Feb 2016 1 Ways to measure relatedness 2 Pedigree-based kinship coefficients
Kinship/relatedness David Balding Professor of Statistical Genetics University of Melbourne, and University College London 2 Feb 2016 1 Ways to measure relatedness 2 Pedigree-based kinship coefficients
Gene coancestry in pedigrees and populations
 Gene coancestry in pedigrees and populations Thompson, Elizabeth University of Washington, Department of Statistics Box 354322 Seattle, WA 98115-4322, USA E-mail: eathomp@uw.edu Glazner, Chris University
Gene coancestry in pedigrees and populations Thompson, Elizabeth University of Washington, Department of Statistics Box 354322 Seattle, WA 98115-4322, USA E-mail: eathomp@uw.edu Glazner, Chris University
Lecture 6: Inbreeding. September 10, 2012
 Lecture 6: Inbreeding September 0, 202 Announcements Hari s New Office Hours Tues 5-6 pm Wed 3-4 pm Fri 2-3 pm In computer lab 3306 LSB Last Time More Hardy-Weinberg Calculations Merle Patterning in Dogs:
Lecture 6: Inbreeding September 0, 202 Announcements Hari s New Office Hours Tues 5-6 pm Wed 3-4 pm Fri 2-3 pm In computer lab 3306 LSB Last Time More Hardy-Weinberg Calculations Merle Patterning in Dogs:
ARTICLE PRIMUS: Rapid Reconstruction of Pedigrees from Genome-wide Estimates of Identity by Descent
 ARTICLE PRIMUS: Rapid Reconstruction of Pedigrees from Genome-wide Estimates of Identity by Descent Jeffrey Staples, 1 Dandi Qiao, 2,3 Michael H. Cho, 2,4 Edwin K. Silverman, 2,4 University of Washington
ARTICLE PRIMUS: Rapid Reconstruction of Pedigrees from Genome-wide Estimates of Identity by Descent Jeffrey Staples, 1 Dandi Qiao, 2,3 Michael H. Cho, 2,4 Edwin K. Silverman, 2,4 University of Washington
Walter Steets Houston Genealogical Forum DNA Interest Group January 6, 2018
 DNA, Ancestry, and Your Genealogical Research- Segments and centimorgans Walter Steets Houston Genealogical Forum DNA Interest Group January 6, 2018 1 Today s agenda Brief review of previous DIG session
DNA, Ancestry, and Your Genealogical Research- Segments and centimorgans Walter Steets Houston Genealogical Forum DNA Interest Group January 6, 2018 1 Today s agenda Brief review of previous DIG session
From: Prof. Carlos D. Bustamante, Ph.D. Date: October 10, 2018
 From: Prof. Carlos D. Bustamante, Ph.D. Date: October 10, 2018 Executive Summary. We find strong evidence that a DNA sample of primarily European descent also contains Native American ancestry from an
From: Prof. Carlos D. Bustamante, Ph.D. Date: October 10, 2018 Executive Summary. We find strong evidence that a DNA sample of primarily European descent also contains Native American ancestry from an
Kenneth Nordtvedt. Many genetic genealogists eventually employ a time-tomost-recent-common-ancestor
 Kenneth Nordtvedt Many genetic genealogists eventually employ a time-tomost-recent-common-ancestor (TMRCA) tool to estimate how far back in time the common ancestor existed for two Y-STR haplotypes obtained
Kenneth Nordtvedt Many genetic genealogists eventually employ a time-tomost-recent-common-ancestor (TMRCA) tool to estimate how far back in time the common ancestor existed for two Y-STR haplotypes obtained
Detecting Heterogeneity in Population Structure Across the Genome in Admixed Populations
 Genetics: Early Online, published on July 20, 2016 as 10.1534/genetics.115.184184 GENETICS INVESTIGATION Detecting Heterogeneity in Population Structure Across the Genome in Admixed Populations Caitlin
Genetics: Early Online, published on July 20, 2016 as 10.1534/genetics.115.184184 GENETICS INVESTIGATION Detecting Heterogeneity in Population Structure Across the Genome in Admixed Populations Caitlin
DNA: UNLOCKING THE CODE
 DNA: UNLOCKING THE CODE Connecting Cousins for Genetic Genealogy Bryant McAllister, PhD Associate Professor of Biology University of Iowa bryant-mcallister@uiowa.edu Iowa Genealogical Society April 9,
DNA: UNLOCKING THE CODE Connecting Cousins for Genetic Genealogy Bryant McAllister, PhD Associate Professor of Biology University of Iowa bryant-mcallister@uiowa.edu Iowa Genealogical Society April 9,
Genetic Research in Utah
 Genetic Research in Utah Lisa Cannon Albright, PhD Professor, Program Leader Genetic Epidemiology Department of Internal Medicine University of Utah School of Medicine George E. Wahlen Department of Veterans
Genetic Research in Utah Lisa Cannon Albright, PhD Professor, Program Leader Genetic Epidemiology Department of Internal Medicine University of Utah School of Medicine George E. Wahlen Department of Veterans
Genetics: Early Online, published on June 29, 2016 as /genetics A Genealogical Look at Shared Ancestry on the X Chromosome
 Genetics: Early Online, published on June 29, 2016 as 10.1534/genetics.116.190041 GENETICS INVESTIGATION A Genealogical Look at Shared Ancestry on the X Chromosome Vince Buffalo,,1, Stephen M. Mount and
Genetics: Early Online, published on June 29, 2016 as 10.1534/genetics.116.190041 GENETICS INVESTIGATION A Genealogical Look at Shared Ancestry on the X Chromosome Vince Buffalo,,1, Stephen M. Mount and
BIOL 502 Population Genetics Spring 2017
 BIOL 502 Population Genetics Spring 2017 Week 8 Inbreeding Arun Sethuraman California State University San Marcos Table of contents 1. Inbreeding Coefficient 2. Mating Systems 3. Consanguinity and Inbreeding
BIOL 502 Population Genetics Spring 2017 Week 8 Inbreeding Arun Sethuraman California State University San Marcos Table of contents 1. Inbreeding Coefficient 2. Mating Systems 3. Consanguinity and Inbreeding
Ancestral Recombination Graphs
 Ancestral Recombination Graphs Ancestral relationships among a sample of recombining sequences usually cannot be accurately described by just a single genealogy. Linked sites will have similar, but not
Ancestral Recombination Graphs Ancestral relationships among a sample of recombining sequences usually cannot be accurately described by just a single genealogy. Linked sites will have similar, but not
Pedigree Reconstruction using Identity by Descent
 Pedigree Reconstruction using Identity by Descent Bonnie Kirkpatrick Electrical Engineering and Computer Sciences University of California at Berkeley Technical Report No. UCB/EECS-2010-43 http://www.eecs.berkeley.edu/pubs/techrpts/2010/eecs-2010-43.html
Pedigree Reconstruction using Identity by Descent Bonnie Kirkpatrick Electrical Engineering and Computer Sciences University of California at Berkeley Technical Report No. UCB/EECS-2010-43 http://www.eecs.berkeley.edu/pubs/techrpts/2010/eecs-2010-43.html
Genome-Wide Association Exercise - Data Quality Control
 Genome-Wide Association Exercise - Data Quality Control The Rockefeller University, New York, June 25, 2016 Copyright 2016 Merry-Lynn McDonald & Suzanne M. Leal Introduction In this exercise, you will
Genome-Wide Association Exercise - Data Quality Control The Rockefeller University, New York, June 25, 2016 Copyright 2016 Merry-Lynn McDonald & Suzanne M. Leal Introduction In this exercise, you will
Genomic Variation of Inbreeding and Ancestry in the Remaining Two Isle Royale Wolves
 Journal of Heredity, 17, 1 16 doi:1.19/jhered/esw8 Original Article Advance Access publication December 1, 16 Original Article Genomic Variation of Inbreeding and Ancestry in the Remaining Two Isle Royale
Journal of Heredity, 17, 1 16 doi:1.19/jhered/esw8 Original Article Advance Access publication December 1, 16 Original Article Genomic Variation of Inbreeding and Ancestry in the Remaining Two Isle Royale
NIH Public Access Author Manuscript Genet Res (Camb). Author manuscript; available in PMC 2011 April 4.
 NIH Public Access Author Manuscript Published in final edited form as: Genet Res (Camb). 2011 February ; 93(1): 47 64. doi:10.1017/s0016672310000480. Variation in actual relationship as a consequence of
NIH Public Access Author Manuscript Published in final edited form as: Genet Res (Camb). 2011 February ; 93(1): 47 64. doi:10.1017/s0016672310000480. Variation in actual relationship as a consequence of
An Optimal Algorithm for Automatic Genotype Elimination
 Am. J. Hum. Genet. 65:1733 1740, 1999 An Optimal Algorithm for Automatic Genotype Elimination Jeffrey R. O Connell 1,2 and Daniel E. Weeks 1 1 Department of Human Genetics, University of Pittsburgh, Pittsburgh,
Am. J. Hum. Genet. 65:1733 1740, 1999 An Optimal Algorithm for Automatic Genotype Elimination Jeffrey R. O Connell 1,2 and Daniel E. Weeks 1 1 Department of Human Genetics, University of Pittsburgh, Pittsburgh,
Identification of the Hypothesized African Ancestry of the Wife of Pvt. Henry Windecker Using Genomic Testing of the Autosomes.
 Identification of the Hypothesized African Ancestry of the Wife of Pvt. Henry Windecker Using Genomic Testing of the Autosomes Introduction African Ancestry: The hypothesis, based on considerable circumstantial
Identification of the Hypothesized African Ancestry of the Wife of Pvt. Henry Windecker Using Genomic Testing of the Autosomes Introduction African Ancestry: The hypothesis, based on considerable circumstantial
Pedigree Reconstruction Using Identity by Descent
 Pedigree Reconstruction Using Identity by Descent Bonnie Kirkpatrick 1, Shuai Cheng Li 2, Richard M. Karp 3, and Eran Halperin 4 1 Electrical Engineering and Computer Sciences, University of California,
Pedigree Reconstruction Using Identity by Descent Bonnie Kirkpatrick 1, Shuai Cheng Li 2, Richard M. Karp 3, and Eran Halperin 4 1 Electrical Engineering and Computer Sciences, University of California,
Statistical methods in genetic relatedness and pedigree analysis
 Statistical methods in genetic relatedness and pedigree analysis Oslo, January 2018 Magnus Dehli Vigeland and Thore Egeland Exercise set III: Coecients of pairwise relatedness Exercise III-1. Use Wright's
Statistical methods in genetic relatedness and pedigree analysis Oslo, January 2018 Magnus Dehli Vigeland and Thore Egeland Exercise set III: Coecients of pairwise relatedness Exercise III-1. Use Wright's
Inbreeding and self-fertilization
 Inbreeding and self-fertilization Introduction Remember that long list of assumptions associated with derivation of the Hardy-Weinberg principle that we just finished? Well, we re about to begin violating
Inbreeding and self-fertilization Introduction Remember that long list of assumptions associated with derivation of the Hardy-Weinberg principle that we just finished? Well, we re about to begin violating
Figure S5 PCA of individuals run on the EAS array reporting Pacific Islander ethnicity, including those reporting another ethnicity.
 Figure S1 PCA of European and West Asian subjects on the EUR array. A clear Ashkenazi cluster is observed. The largest cluster depicts the northwest southeast cline within Europe. A Those reporting a single
Figure S1 PCA of European and West Asian subjects on the EUR array. A clear Ashkenazi cluster is observed. The largest cluster depicts the northwest southeast cline within Europe. A Those reporting a single
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1122655/dc1 Supporting Online Material for Finding Criminals Through DNA of Their Relatives Frederick R. Bieber,* Charles H. Brenner, David Lazer *Author for correspondence.
www.sciencemag.org/cgi/content/full/1122655/dc1 Supporting Online Material for Finding Criminals Through DNA of Their Relatives Frederick R. Bieber,* Charles H. Brenner, David Lazer *Author for correspondence.
Nature Genetics: doi: /ng Supplementary Figure 1. Quality control of FALS discovery cohort.
 Supplementary Figure 1 Quality control of FALS discovery cohort. Exome sequences were obtained for 1,376 FALS cases and 13,883 controls. Samples were excluded in the event of exome-wide call rate
Supplementary Figure 1 Quality control of FALS discovery cohort. Exome sequences were obtained for 1,376 FALS cases and 13,883 controls. Samples were excluded in the event of exome-wide call rate
On identification problems requiring linked autosomal markers
 * Title Page (with authors & addresses) On identification problems requiring linked autosomal markers Thore Egeland a Nuala Sheehan b a Department of Medical Genetics, Ulleval University Hospital, 0407
* Title Page (with authors & addresses) On identification problems requiring linked autosomal markers Thore Egeland a Nuala Sheehan b a Department of Medical Genetics, Ulleval University Hospital, 0407
NON-RANDOM MATING AND INBREEDING
 Instructor: Dr. Martha B. Reiskind AEC 495/AEC592: Conservation Genetics DEFINITIONS Nonrandom mating: Mating individuals are more closely related or less closely related than those drawn by chance from
Instructor: Dr. Martha B. Reiskind AEC 495/AEC592: Conservation Genetics DEFINITIONS Nonrandom mating: Mating individuals are more closely related or less closely related than those drawn by chance from
Bottlenecks reduce genetic variation Genetic Drift
 Bottlenecks reduce genetic variation Genetic Drift Northern Elephant Seals were reduced to ~30 individuals in the 1800s. Rare alleles are likely to be lost during a bottleneck Two important determinants
Bottlenecks reduce genetic variation Genetic Drift Northern Elephant Seals were reduced to ~30 individuals in the 1800s. Rare alleles are likely to be lost during a bottleneck Two important determinants
Kinship and Population Subdivision
 Kinship and Population Subdivision Henry Harpending University of Utah The coefficient of kinship between two diploid organisms describes their overall genetic similarity to each other relative to some
Kinship and Population Subdivision Henry Harpending University of Utah The coefficient of kinship between two diploid organisms describes their overall genetic similarity to each other relative to some
Chapter 2: Genes in Pedigrees
 Chapter 2: Genes in Pedigrees Chapter 2-0 2.1 Pedigree definitions and terminology 2-1 2.2 Gene identity by descent (ibd) 2-5 2.3 ibd of more than 2 genes 2-14 2.4 Data on relatives 2-21 2.1.1 GRAPHICAL
Chapter 2: Genes in Pedigrees Chapter 2-0 2.1 Pedigree definitions and terminology 2-1 2.2 Gene identity by descent (ibd) 2-5 2.3 ibd of more than 2 genes 2-14 2.4 Data on relatives 2-21 2.1.1 GRAPHICAL
Advanced Autosomal DNA Techniques used in Genetic Genealogy
 Advanced Autosomal DNA Techniques used in Genetic Genealogy Tim Janzen, MD E-mail: tjanzen@comcast.net Summary of Chromosome Mapping Technique The following are specific instructions on how to map your
Advanced Autosomal DNA Techniques used in Genetic Genealogy Tim Janzen, MD E-mail: tjanzen@comcast.net Summary of Chromosome Mapping Technique The following are specific instructions on how to map your
Linkage Analysis in Merlin. Meike Bartels Kate Morley Danielle Posthuma
 Linkage Analysis in Merlin Meike Bartels Kate Morley Danielle Posthuma Software for linkage analyses Genehunter Mendel Vitesse Allegro Simwalk Loki Merlin. Mx R Lisrel MERLIN software Programs: MERLIN
Linkage Analysis in Merlin Meike Bartels Kate Morley Danielle Posthuma Software for linkage analyses Genehunter Mendel Vitesse Allegro Simwalk Loki Merlin. Mx R Lisrel MERLIN software Programs: MERLIN
AFDAA 2012 WINTER MEETING Population Statistics Refresher Course - Lecture 3: Statistics of Kinship Analysis
 AFDAA 2012 WINTER MEETING Population Statistics Refresher Course - Lecture 3: Statistics of Kinship Analysis Ranajit Chakraborty, PhD Center for Computational Genomics Institute of Applied Genetics Department
AFDAA 2012 WINTER MEETING Population Statistics Refresher Course - Lecture 3: Statistics of Kinship Analysis Ranajit Chakraborty, PhD Center for Computational Genomics Institute of Applied Genetics Department
2 The Wright-Fisher model and the neutral theory
 0 THE WRIGHT-FISHER MODEL AND THE NEUTRAL THEORY The Wright-Fisher model and the neutral theory Although the main interest of population genetics is conceivably in natural selection, we will first assume
0 THE WRIGHT-FISHER MODEL AND THE NEUTRAL THEORY The Wright-Fisher model and the neutral theory Although the main interest of population genetics is conceivably in natural selection, we will first assume
Inbreeding and self-fertilization
 Inbreeding and self-fertilization Introduction Remember that long list of assumptions associated with derivation of the Hardy-Weinberg principle that I went over a couple of lectures ago? Well, we re about
Inbreeding and self-fertilization Introduction Remember that long list of assumptions associated with derivation of the Hardy-Weinberg principle that I went over a couple of lectures ago? Well, we re about
Population Structure. Population Structure
 Nonrandom Mating HWE assumes that mating is random in the population Most natural populations deviate in some way from random mating There are various ways in which a species might deviate from random
Nonrandom Mating HWE assumes that mating is random in the population Most natural populations deviate in some way from random mating There are various ways in which a species might deviate from random
Autosomal DNA. What is autosomal DNA? X-DNA
 ANGIE BUSH AND PAUL WOODBURY info@thednadetectives.com November 1, 2014 Autosomal DNA What is autosomal DNA? Autosomal DNA consists of all nuclear DNA except for the X and Y sex chromosomes. There are
ANGIE BUSH AND PAUL WOODBURY info@thednadetectives.com November 1, 2014 Autosomal DNA What is autosomal DNA? Autosomal DNA consists of all nuclear DNA except for the X and Y sex chromosomes. There are
4. Kinship Paper Challenge
 4. António Amorim (aamorim@ipatimup.pt) Nádia Pinto (npinto@ipatimup.pt) 4.1 Approach After a woman dies her child claims for a paternity test of the man who is supposed to be his father. The test is carried
4. António Amorim (aamorim@ipatimup.pt) Nádia Pinto (npinto@ipatimup.pt) 4.1 Approach After a woman dies her child claims for a paternity test of the man who is supposed to be his father. The test is carried
Optimum contribution selection conserves genetic diversity better than random selection in small populations with overlapping generations
 Optimum contribution selection conserves genetic diversity better than random selection in small populations with overlapping generations K. Stachowicz 12*, A. C. Sørensen 23 and P. Berg 3 1 Department
Optimum contribution selection conserves genetic diversity better than random selection in small populations with overlapping generations K. Stachowicz 12*, A. C. Sørensen 23 and P. Berg 3 1 Department
Methods of Parentage Analysis in Natural Populations
 Methods of Parentage Analysis in Natural Populations Using molecular markers, estimates of genetic maternity or paternity can be achieved by excluding as parents all adults whose genotypes are incompatible
Methods of Parentage Analysis in Natural Populations Using molecular markers, estimates of genetic maternity or paternity can be achieved by excluding as parents all adults whose genotypes are incompatible
Comparative method, coalescents, and the future. Correlation of states in a discrete-state model
 Comparative method, coalescents, and the future Joe Felsenstein Depts. of Genome Sciences and of Biology, University of Washington Comparative method, coalescents, and the future p.1/28 Correlation of
Comparative method, coalescents, and the future Joe Felsenstein Depts. of Genome Sciences and of Biology, University of Washington Comparative method, coalescents, and the future p.1/28 Correlation of
DNA Testing. February 16, 2018
 DNA Testing February 16, 2018 What Is DNA? Double helix ladder structure where the rungs are molecules called nucleotides or bases. DNA contains only four of these nucleotides A, G, C, T The sequence that
DNA Testing February 16, 2018 What Is DNA? Double helix ladder structure where the rungs are molecules called nucleotides or bases. DNA contains only four of these nucleotides A, G, C, T The sequence that
Supplementary Note: Analysis of Latino populations from GALA and MEC reveals genomic loci with biased local ancestry estimation
 Supplementary Note: Analysis of Latino populations from GALA and MEC reveals genomic loci with biased local ancestry estimation Bogdan Pasaniuc, Sriram Sankararaman, et al. 1 Relation between Error Rate
Supplementary Note: Analysis of Latino populations from GALA and MEC reveals genomic loci with biased local ancestry estimation Bogdan Pasaniuc, Sriram Sankararaman, et al. 1 Relation between Error Rate
Genetic Genealogy. Rules and Tools. Baltimore County Genealogical Society March 25, 2018 Andrew Hochreiter
 Genetic Genealogy Rules and Tools Baltimore County Genealogical Society March 25, 2018 Andrew Hochreiter I am NOT this guy! 2 Genealogy s Newest Tool Genealogy research: Study of Family History Identifies
Genetic Genealogy Rules and Tools Baltimore County Genealogical Society March 25, 2018 Andrew Hochreiter I am NOT this guy! 2 Genealogy s Newest Tool Genealogy research: Study of Family History Identifies
Objective: Why? 4/6/2014. Outlines:
 Objective: Develop mathematical models that quantify/model resemblance between relatives for phenotypes of a quantitative trait : - based on pedigree - based on markers Outlines: Causal model for covariances
Objective: Develop mathematical models that quantify/model resemblance between relatives for phenotypes of a quantitative trait : - based on pedigree - based on markers Outlines: Causal model for covariances
CAGGNI s DNA Special Interest Group
 CAGGNI s DNA Special Interest Group 10 Jan 2015 Al & Michelle Wilson Agenda Survey Basics in Fan Charts Recombination Exercise Triangulation Overview Survey 1. Have you taken (or sponsored) a DNA test?
CAGGNI s DNA Special Interest Group 10 Jan 2015 Al & Michelle Wilson Agenda Survey Basics in Fan Charts Recombination Exercise Triangulation Overview Survey 1. Have you taken (or sponsored) a DNA test?
Genealogical Research
 DNA, Ancestry, and Your Genealogical Research Walter Steets Houston Genealogical Forum DNA Interest Group March 2, 2019 1 Today s Agenda Brief review of basic genetics and terms used in genetic genealogy
DNA, Ancestry, and Your Genealogical Research Walter Steets Houston Genealogical Forum DNA Interest Group March 2, 2019 1 Today s Agenda Brief review of basic genetics and terms used in genetic genealogy
Comparative method, coalescents, and the future
 Comparative method, coalescents, and the future Joe Felsenstein Depts. of Genome Sciences and of Biology, University of Washington Comparative method, coalescents, and the future p.1/36 Correlation of
Comparative method, coalescents, and the future Joe Felsenstein Depts. of Genome Sciences and of Biology, University of Washington Comparative method, coalescents, and the future p.1/36 Correlation of
CONGEN. Inbreeding vocabulary
 CONGEN Inbreeding vocabulary Inbreeding Mating between relatives. Inbreeding depression Reduction in fitness due to inbreeding. Identical by descent Alleles that are identical by descent are direct descendents
CONGEN Inbreeding vocabulary Inbreeding Mating between relatives. Inbreeding depression Reduction in fitness due to inbreeding. Identical by descent Alleles that are identical by descent are direct descendents
Factors affecting phasing quality in a commercial layer population
 Factors affecting phasing quality in a commercial layer population N. Frioni 1, D. Cavero 2, H. Simianer 1 & M. Erbe 3 1 University of Goettingen, Department of nimal Sciences, Center for Integrated Breeding
Factors affecting phasing quality in a commercial layer population N. Frioni 1, D. Cavero 2, H. Simianer 1 & M. Erbe 3 1 University of Goettingen, Department of nimal Sciences, Center for Integrated Breeding
Lecture 1: Introduction to pedigree analysis
 Lecture 1: Introduction to pedigree analysis Magnus Dehli Vigeland NORBIS course, 8 th 12 th of January 2018, Oslo Outline Part I: Brief introductions Pedigrees symbols and terminology Some common relationships
Lecture 1: Introduction to pedigree analysis Magnus Dehli Vigeland NORBIS course, 8 th 12 th of January 2018, Oslo Outline Part I: Brief introductions Pedigrees symbols and terminology Some common relationships
Algorithms for Genetics: Basics of Wright Fisher Model and Coalescent Theory
 Algorithms for Genetics: Basics of Wright Fisher Model and Coalescent Theory Vineet Bafna Harish Nagarajan and Nitin Udpa 1 Disclaimer Please note that a lot of the text and figures here are copied from
Algorithms for Genetics: Basics of Wright Fisher Model and Coalescent Theory Vineet Bafna Harish Nagarajan and Nitin Udpa 1 Disclaimer Please note that a lot of the text and figures here are copied from
Inbreeding Using Genomics and How it Can Help. Dr. Flavio S. Schenkel CGIL- University of Guelph
 Inbreeding Using Genomics and How it Can Help Dr. Flavio S. Schenkel CGIL- University of Guelph Introduction Why is inbreeding a concern? The biological risks of inbreeding: Inbreeding depression Accumulation
Inbreeding Using Genomics and How it Can Help Dr. Flavio S. Schenkel CGIL- University of Guelph Introduction Why is inbreeding a concern? The biological risks of inbreeding: Inbreeding depression Accumulation
Growing the Family Tree: The Power of DNA in Reconstructing Family Relationships
 Growing the Family Tree: The Power of DNA in Reconstructing Family Relationships Luke A. D. Hutchison Natalie M. Myres Scott R. Woodward Sorenson Molecular Genealogy Foundation (www.smgf.org) 2511 South
Growing the Family Tree: The Power of DNA in Reconstructing Family Relationships Luke A. D. Hutchison Natalie M. Myres Scott R. Woodward Sorenson Molecular Genealogy Foundation (www.smgf.org) 2511 South
Coalescence. Outline History. History, Model, and Application. Coalescence. The Model. Application
 Coalescence History, Model, and Application Outline History Origins of theory/approach Trace the incorporation of other s ideas Coalescence Definition and descriptions The Model Assumptions and Uses Application
Coalescence History, Model, and Application Outline History Origins of theory/approach Trace the incorporation of other s ideas Coalescence Definition and descriptions The Model Assumptions and Uses Application
[CLIENT] SmithDNA1701 DE January 2017
![[CLIENT] SmithDNA1701 DE January 2017 [CLIENT] SmithDNA1701 DE January 2017](/thumbs/93/113058427.jpg) [CLIENT] SmithDNA1701 DE1704205 11 January 2017 DNA Discovery Plan GOAL Create a research plan to determine how the client s DNA results relate to his family tree as currently constructed. The client s
[CLIENT] SmithDNA1701 DE1704205 11 January 2017 DNA Discovery Plan GOAL Create a research plan to determine how the client s DNA results relate to his family tree as currently constructed. The client s
Assessment of alternative genotyping strategies to maximize imputation accuracy at minimal cost
 Huang et al. Genetics Selection Evolution 2012, 44:25 Genetics Selection Evolution RESEARCH Open Access Assessment of alternative genotyping strategies to maximize imputation accuracy at minimal cost Yijian
Huang et al. Genetics Selection Evolution 2012, 44:25 Genetics Selection Evolution RESEARCH Open Access Assessment of alternative genotyping strategies to maximize imputation accuracy at minimal cost Yijian
Forensic use of the genomic relationship matrix to validate and discover livestock. pedigrees
 Forensic use of the genomic relationship matrix to validate and discover livestock pedigrees K. L. Moore*, C. Vilela*, K. Kaseja*, R, Mrode* and M. Coffey* * Scotland s Rural College (SRUC), Easter Bush,
Forensic use of the genomic relationship matrix to validate and discover livestock pedigrees K. L. Moore*, C. Vilela*, K. Kaseja*, R, Mrode* and M. Coffey* * Scotland s Rural College (SRUC), Easter Bush,
Large scale kinship:familial Searching and DVI. Seoul, ISFG workshop
 Large scale kinship:familial Searching and DVI Seoul, ISFG workshop 29 August 2017 Large scale kinship Familial Searching: search for a relative of an unidentified offender whose profile is available in
Large scale kinship:familial Searching and DVI Seoul, ISFG workshop 29 August 2017 Large scale kinship Familial Searching: search for a relative of an unidentified offender whose profile is available in
Population Structure and Genealogies
 Population Structure and Genealogies One of the key properties of Kingman s coalescent is that each pair of lineages is equally likely to coalesce whenever a coalescent event occurs. This condition is
Population Structure and Genealogies One of the key properties of Kingman s coalescent is that each pair of lineages is equally likely to coalesce whenever a coalescent event occurs. This condition is
Walter Steets Houston Genealogical Forum DNA Interest Group April 7, 2018
 Ancestry DNA and GEDmatch Walter Steets Houston Genealogical Forum DNA Interest Group April 7, 2018 Today s agenda Recent News about DNA Testing DNA Cautions: DNA Data Used for Forensic Purposes New Technology:
Ancestry DNA and GEDmatch Walter Steets Houston Genealogical Forum DNA Interest Group April 7, 2018 Today s agenda Recent News about DNA Testing DNA Cautions: DNA Data Used for Forensic Purposes New Technology:
! FTDNA! Ancestry. ! 23andMe. ! Medical Considera,ons. ! Iden,fying family medical history. ! Communica,ng with the medical community
 by JEFF CARPENTER! Brief Defini,ons about YDNA, XDNA, mtdna, atdna (Covered in Part 1)! Benefits of Tes,ng DNA! Examples of DNA TESTING! FTDNA! Ancestry! 3andMe Jeff Carpenter, 016 jeffcarpenter1939@gmal.com!
by JEFF CARPENTER! Brief Defini,ons about YDNA, XDNA, mtdna, atdna (Covered in Part 1)! Benefits of Tes,ng DNA! Examples of DNA TESTING! FTDNA! Ancestry! 3andMe Jeff Carpenter, 016 jeffcarpenter1939@gmal.com!
Introduction. Article 50 million: an estimate of the number of scholarly articles in existence RESEARCH ARTICLE
 Article 50 million: an estimate of the number of scholarly articles in existence Arif E. Jinha 258 Arif E. Jinha Learned Publishing, 23:258 263 doi:10.1087/20100308 Arif E. Jinha Introduction From the
Article 50 million: an estimate of the number of scholarly articles in existence Arif E. Jinha 258 Arif E. Jinha Learned Publishing, 23:258 263 doi:10.1087/20100308 Arif E. Jinha Introduction From the
Introduction to Autosomal DNA Tools
 GENETIC GENEALOGY JOURNEY Debbie Parker Wayne, CG, CGL Introduction to Autosomal DNA Tools Just as in the old joke about a new genealogist walking into the library and asking for the book that covers my
GENETIC GENEALOGY JOURNEY Debbie Parker Wayne, CG, CGL Introduction to Autosomal DNA Tools Just as in the old joke about a new genealogist walking into the library and asking for the book that covers my
KinLinks: Software Toolkit for Kinship Analysis and Pedigree Generation from NGS Datasets
 KinLinks: Software Toolkit for Kinship Analysis and Pedigree Generation from NGS Datasets Anna Shcherbina*, Darrell Ricke, Eric Schwoebel, Tara Boettcher, Christina Zook, Johanna Bobrow, Martha Petrovick,
KinLinks: Software Toolkit for Kinship Analysis and Pedigree Generation from NGS Datasets Anna Shcherbina*, Darrell Ricke, Eric Schwoebel, Tara Boettcher, Christina Zook, Johanna Bobrow, Martha Petrovick,
Using Y-DNA for Genealogy Debbie Parker Wayne, CG, CGL SM
 Using Y-DNA for Genealogy Debbie Parker Wayne, CG, CGL SM This is one article of a series on using DNA for genealogical research. There are several types of DNA tests offered for genealogical purposes.
Using Y-DNA for Genealogy Debbie Parker Wayne, CG, CGL SM This is one article of a series on using DNA for genealogical research. There are several types of DNA tests offered for genealogical purposes.
DNA Basics, Y DNA Marker Tables, Ancestral Trees and Mutation Graphs: Definitions, Concepts, Understanding
 DNA Basics, Y DNA Marker Tables, Ancestral Trees and Mutation Graphs: Definitions, Concepts, Understanding by Dr. Ing. Robert L. Baber 2014 July 26 Rights reserved, see the copyright notice at http://gengen.rlbaber.de
DNA Basics, Y DNA Marker Tables, Ancestral Trees and Mutation Graphs: Definitions, Concepts, Understanding by Dr. Ing. Robert L. Baber 2014 July 26 Rights reserved, see the copyright notice at http://gengen.rlbaber.de
Recent effective population size estimated from segments of identity by descent in the Lithuanian population
 Anthropological Science Advance Publication Recent effective population size estimated from segments of identity by descent in the Lithuanian population Alina Urnikytė 1 *, Alma Molytė 1, Vaidutis Kučinskas
Anthropological Science Advance Publication Recent effective population size estimated from segments of identity by descent in the Lithuanian population Alina Urnikytė 1 *, Alma Molytė 1, Vaidutis Kučinskas
TRACK 1: BEGINNING DNA RESEARCH presented by Andy Hochreiter
 TRACK 1: BEGINNING DNA RESEARCH presented by Andy Hochreiter 1-1: DNA: WHERE DO I START? Definition Genetic genealogy is the application of genetics to traditional genealogy. Genetic genealogy uses genealogical
TRACK 1: BEGINNING DNA RESEARCH presented by Andy Hochreiter 1-1: DNA: WHERE DO I START? Definition Genetic genealogy is the application of genetics to traditional genealogy. Genetic genealogy uses genealogical
Discovering Hard to Find Ancestry DNA Matches Page 1
 Discovering Hard To Find Ancestry DNA Matches Alice Kalush 5/15/2018 This document discusses several methods for finding matches to your Ancestry DNA test that do not easily show up for you in the Hints
Discovering Hard To Find Ancestry DNA Matches Alice Kalush 5/15/2018 This document discusses several methods for finding matches to your Ancestry DNA test that do not easily show up for you in the Hints
DNA Testing What you need to know first
 DNA Testing What you need to know first This article is like the Cliff Notes version of several genetic genealogy classes. It is a basic general primer. The general areas include Project support DNA test
DNA Testing What you need to know first This article is like the Cliff Notes version of several genetic genealogy classes. It is a basic general primer. The general areas include Project support DNA test
Walter Steets Houston Genealogical Forum DNA Interest Group February 24, 2018
 Using Ancestry DNA and Third-Party Tools to Research Your Shared DNA Segments Part 2 Walter Steets Houston Genealogical Forum DNA Interest Group February 24, 2018 1 Today s agenda Brief review of previous
Using Ancestry DNA and Third-Party Tools to Research Your Shared DNA Segments Part 2 Walter Steets Houston Genealogical Forum DNA Interest Group February 24, 2018 1 Today s agenda Brief review of previous
LASER server: ancestry tracing with genotypes or sequence reads
 LASER server: ancestry tracing with genotypes or sequence reads The LASER method Supplementary Data For each ancestry reference panel of N individuals, LASER applies principal components analysis (PCA)
LASER server: ancestry tracing with genotypes or sequence reads The LASER method Supplementary Data For each ancestry reference panel of N individuals, LASER applies principal components analysis (PCA)
CLOSE relatives are expected to share large contiguous. A Genealogical Look at Shared Ancestry on the X Chromosome INVESTIGATION
 INVESTIGATION A Genealogical Look at Shared Ancestry on the X Chromosome Vince Buffalo,*,,1 Stephen M. Mount, and Graham Coop *Population Biology Graduate Group, Center for Population Biology, Department
INVESTIGATION A Genealogical Look at Shared Ancestry on the X Chromosome Vince Buffalo,*,,1 Stephen M. Mount, and Graham Coop *Population Biology Graduate Group, Center for Population Biology, Department
Tools: 23andMe.com website and test results; DNAAdoption handouts.
 When You First Get Your 23andMe Results Objective: Learn what to do with results of atdna testing with 23andMe. Tools: 23andMe.com website and test results; DNAAdoption handouts. Exercises: Practice Exercises
When You First Get Your 23andMe Results Objective: Learn what to do with results of atdna testing with 23andMe. Tools: 23andMe.com website and test results; DNAAdoption handouts. Exercises: Practice Exercises
Edinburgh Research Explorer
 Edinburgh Research Explorer Runs of Homozygosity in European Populations Citation for published version: McQuillan, R, Leutenegger, A-L, Abdel-Rahman, R, Franklin, CS, Pericic, M, Barac-Lauc, L, Smolej-
Edinburgh Research Explorer Runs of Homozygosity in European Populations Citation for published version: McQuillan, R, Leutenegger, A-L, Abdel-Rahman, R, Franklin, CS, Pericic, M, Barac-Lauc, L, Smolej-
Using Autosomal DNA for Genealogy Debbie Parker Wayne, CG, CGL SM
 Using Autosomal DNA for Genealogy Debbie Parker Wayne, CG, CGL SM This is one article of a series on using DNA for genealogical research. There are several types of DNA tests offered for genealogical purposes.
Using Autosomal DNA for Genealogy Debbie Parker Wayne, CG, CGL SM This is one article of a series on using DNA for genealogical research. There are several types of DNA tests offered for genealogical purposes.
Puzzling Pedigrees. Essential Question: How can pedigrees be used to study the inheritance of human traits?
 Name: Puzzling Pedigrees Essential Question: How can pedigrees be used to study the inheritance of human traits? Studying inheritance in humans is more difficult than studying inheritance in fruit flies
Name: Puzzling Pedigrees Essential Question: How can pedigrees be used to study the inheritance of human traits? Studying inheritance in humans is more difficult than studying inheritance in fruit flies
Some of these slides have been borrowed from Dr. Paul Lewis, Dr. Joe Felsenstein. Thanks!
 Some of these slides have been borrowed from Dr. Paul Lewis, Dr. Joe Felsenstein. Thanks! Paul has many great tools for teaching phylogenetics at his web site: http://hydrodictyon.eeb.uconn.edu/people/plewis
Some of these slides have been borrowed from Dr. Paul Lewis, Dr. Joe Felsenstein. Thanks! Paul has many great tools for teaching phylogenetics at his web site: http://hydrodictyon.eeb.uconn.edu/people/plewis
Forensic Statistics and Graphical Models (1) Richard Gill Spring Semester 2015
 Forensic Statistics and Graphical Models (1) Richard Gill Spring Semester 2015 http://www.math.leidenuniv.nl/~gill/teaching/graphical Forensic Statistics Distinguish criminal investigation and criminal
Forensic Statistics and Graphical Models (1) Richard Gill Spring Semester 2015 http://www.math.leidenuniv.nl/~gill/teaching/graphical Forensic Statistics Distinguish criminal investigation and criminal
1 NOTE: This paper reports the results of research and analysis
 Race and Hispanic Origin Data: A Comparison of Results From the Census 2000 Supplementary Survey and Census 2000 Claudette E. Bennett and Deborah H. Griffin, U. S. Census Bureau Claudette E. Bennett, U.S.
Race and Hispanic Origin Data: A Comparison of Results From the Census 2000 Supplementary Survey and Census 2000 Claudette E. Bennett and Deborah H. Griffin, U. S. Census Bureau Claudette E. Bennett, U.S.
ICMP DNA REPORTS GUIDE
 ICMP DNA REPORTS GUIDE Distribution: General Sarajevo, 16 th December 2010 GUIDE TO ICMP DNA REPORTS 1. Purpose of This Document 1. The International Commission on Missing Persons (ICMP) endeavors to secure
ICMP DNA REPORTS GUIDE Distribution: General Sarajevo, 16 th December 2010 GUIDE TO ICMP DNA REPORTS 1. Purpose of This Document 1. The International Commission on Missing Persons (ICMP) endeavors to secure
BIOL Evolution. Lecture 8
 BIOL 432 - Evolution Lecture 8 Expected Genotype Frequencies in the Absence of Evolution are Determined by the Hardy-Weinberg Equation. Assumptions: 1) No mutation 2) Random mating 3) Infinite population
BIOL 432 - Evolution Lecture 8 Expected Genotype Frequencies in the Absence of Evolution are Determined by the Hardy-Weinberg Equation. Assumptions: 1) No mutation 2) Random mating 3) Infinite population
The genealogical history of a population The coalescent process. Identity by descent Distribution of pairwise coalescence times
 The coalescent The genealogical history of a population The coalescent process Identity by descent Distribution of pairwise coalescence times Adding mutations Expected pairwise differences Evolutionary
The coalescent The genealogical history of a population The coalescent process Identity by descent Distribution of pairwise coalescence times Adding mutations Expected pairwise differences Evolutionary
Walter Steets Houston Genealogical Forum DNA Interest Group November 18, 2017
 DNA, Ancestry, and Your Genealogical Research Session 2 Walter Steets Houston Genealogical Forum DNA Interest Group November 18, 2017 1 Today s agenda Brief review of previous DIG session Degrees of Separation
DNA, Ancestry, and Your Genealogical Research Session 2 Walter Steets Houston Genealogical Forum DNA Interest Group November 18, 2017 1 Today s agenda Brief review of previous DIG session Degrees of Separation
GEDmatch Home Page The upper left corner of your home page has Information about you and links to lots of helpful information. Check them out!
 USING GEDMATCH Created March 2015 GEDmatch is a free, non-profit site that accepts raw autosomal data files from Ancestry, FTDNA, and 23andme. As such, it provides a large autosomal database that spans
USING GEDMATCH Created March 2015 GEDmatch is a free, non-profit site that accepts raw autosomal data files from Ancestry, FTDNA, and 23andme. As such, it provides a large autosomal database that spans
Big Y-700 White Paper
 Big Y-700 White Paper Powering discovery in the field of paternal ancestry Authors: Caleb Davis, Michael Sager, Göran Runfeldt, Elliott Greenspan, Arjan Bormans, Bennett Greenspan, and Connie Bormans Last
Big Y-700 White Paper Powering discovery in the field of paternal ancestry Authors: Caleb Davis, Michael Sager, Göran Runfeldt, Elliott Greenspan, Arjan Bormans, Bennett Greenspan, and Connie Bormans Last
TDT vignette Use of snpstats in family based studies
 TDT vignette Use of snpstats in family based studies David Clayton April 30, 2018 Pedigree data The snpstats package contains some tools for analysis of family-based studies. These assume that a subject
TDT vignette Use of snpstats in family based studies David Clayton April 30, 2018 Pedigree data The snpstats package contains some tools for analysis of family-based studies. These assume that a subject
Exercise 4 Exploring Population Change without Selection
 Exercise 4 Exploring Population Change without Selection This experiment began with nine Avidian ancestors of identical fitness; the mutation rate is zero percent. Since descendants can never differ in
Exercise 4 Exploring Population Change without Selection This experiment began with nine Avidian ancestors of identical fitness; the mutation rate is zero percent. Since descendants can never differ in
Automated Discovery of Pedigrees and Their Structures in Collections of STR DNA Specimens Using a Link Discovery Tool
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange Masters Theses Graduate School 5-2010 Automated Discovery of Pedigrees and Their Structures in Collections of STR DNA
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange Masters Theses Graduate School 5-2010 Automated Discovery of Pedigrees and Their Structures in Collections of STR DNA
Developing Conclusions About Different Modes of Inheritance
 Pedigree Analysis Introduction A pedigree is a diagram of family relationships that uses symbols to represent people and lines to represent genetic relationships. These diagrams make it easier to visualize
Pedigree Analysis Introduction A pedigree is a diagram of family relationships that uses symbols to represent people and lines to represent genetic relationships. These diagrams make it easier to visualize
TREES OF GENES IN POPULATIONS
 1 TREES OF GENES IN POPULATIONS Joseph Felsenstein Abstract Trees of ancestry of copies of genes form in populations, as a result of the randomness of birth, death, and Mendelian reproduction. Considering
1 TREES OF GENES IN POPULATIONS Joseph Felsenstein Abstract Trees of ancestry of copies of genes form in populations, as a result of the randomness of birth, death, and Mendelian reproduction. Considering
The DNA Case for Bethuel Riggs
 The DNA Case for Bethuel Riggs The following was originally intended as an appendix to Alvy Ray Smith, Edwardian Riggses of America I: Elder Bethuel Riggs (1757 1835) of Morris County, New Jersey, and
The DNA Case for Bethuel Riggs The following was originally intended as an appendix to Alvy Ray Smith, Edwardian Riggses of America I: Elder Bethuel Riggs (1757 1835) of Morris County, New Jersey, and
Forward thinking: the predictive approach
 Coalescent Theory 1 Forward thinking: the predictive approach Random variation in reproduction causes random fluctuation in allele frequencies. Can describe this process as diffusion: (Wright 1931) showed
Coalescent Theory 1 Forward thinking: the predictive approach Random variation in reproduction causes random fluctuation in allele frequencies. Can describe this process as diffusion: (Wright 1931) showed
Analysis of geographically structured populations: Estimators based on coalescence
 Analysis of geographically structured populations: Estimators based on coalescence Peter Beerli Department of Genetics, Box 357360, University of Washington, Seattle WA 9895-7360, Email: beerli@genetics.washington.edu
Analysis of geographically structured populations: Estimators based on coalescence Peter Beerli Department of Genetics, Box 357360, University of Washington, Seattle WA 9895-7360, Email: beerli@genetics.washington.edu
Two-point linkage analysis using the LINKAGE/FASTLINK programs
 1 Two-point linkage analysis using the LINKAGE/FASTLINK programs Copyrighted 2018 Maria Chahrour and Suzanne M. Leal These exercises will introduce the LINKAGE file format which is the standard format
1 Two-point linkage analysis using the LINKAGE/FASTLINK programs Copyrighted 2018 Maria Chahrour and Suzanne M. Leal These exercises will introduce the LINKAGE file format which is the standard format
The History of African Gene Flow into Southern Europeans, Levantines, and Jews
 The History of African Gene Flow into Southern Europeans, Levantines, and Jews Priya Moorjani 1,2 *, Nick Patterson 2, Joel N. Hirschhorn 1,2,3, Alon Keinan 4, Li Hao 5, Gil Atzmon 6, Edward Burns 6, Harry
The History of African Gene Flow into Southern Europeans, Levantines, and Jews Priya Moorjani 1,2 *, Nick Patterson 2, Joel N. Hirschhorn 1,2,3, Alon Keinan 4, Li Hao 5, Gil Atzmon 6, Edward Burns 6, Harry
