Sequencing Method Manual
|
|
|
- Avis Butler
- 6 years ago
- Views:
Transcription
1 Sequencing Method Manual Instrument GS Junior GS FLX+ GS FLX+ GS FLX Kit Junior XL+ XLR70 XLR70 For life science research only. Not for use in diagnostic procedures.
2 1 WORKFLOW GS FLX + Instrument Operation Instrument Preparation This procedure can also be used to perform a test sequencing Run, using only Control DNA Beads. This requires a GS FLX Titanium Control Bead Kit, in addition to all the materials used for an XL+ Run. The only difference is the preparation of the DNA Beads. For details of the preparation of the Control DNA Beads, please see the Package Insert to the GS FLX Titanium Control Bead Kit. PicoTiterPlate Device Preparation The Pre-Wash (section 3.1) Launch the Pre -wash PicoTiterPlate (PTP) Device Preparation (section 3.2) Prepare Bead Buffer 2 (BB2) The Sequencing Run (section 3.3) Remove the Pre-Wash Cassette and Clean the Fluidics Area Deck Prepare the PicoTiterPlate and Bead Deposition Devices Prepare the Beads Prepare and Load the Sequencing Reagents Cassette Clean the PicoTiterPlate Cartridge and the Camera Faceplate Load and Set the Run Script and Other Run Parameters Assemble the BDD with the PTP Device and Bead Loading Gasket Deposit the Four Layers of Beads on the PTP Device Insert the PicoTiterPlate Device and Launch the Sequencing Run Figure 1: Workflow of a sequencing Run performed on the GS FLX+ Instrument, with the GS FLX Titanium Sequencing Kit XL+ Note that instrument preparation and PicoTiterPlate device preparation are done in parallel such that the sequencing Run is initiated as soon as the PicoTiterPlate device is ready. 2/2
3 2 BEFORE YOU BEGIN Room temperature is +15 to +25 C. 2.1 What You Should Have Before Starting Sample The sample library being sequenced must have been prepared using the GS FLX Titanium Series XL+ methods and the corresponding kits Required GS FLX+ System Equipment and Reagents Refer to the customer access area of our web site, at for the lists of materials and consumables required but not provided. This method is for a GS FLX Titanium Sequencing Kit XL+ and a GS FLX+ Instrument Kits The GS FLX Titanium Sequencing Kit XL+ is used in combination with the matching GS FLX Titanium PicoTiterPlate Kit 70 x 75. These kits provide reagents and components necessary for a single sequencing Run. Refer to the customer access area of our web site, at for a complete list of kits and reagents Choosing the Size and Number of PTP Regions for Your Sample The GS FLX Titanium Sequencing Kit XL+ and PicoTiterPlate Kit support various bead loading region configurations. The size and number of PicoTiterPlate (PTP) regions to use depend on the sequencing throughput requirements of the experiment, in total bases or total reads. To determine the proper gasket size, proceed as follows: Use Table 1 and Table 2 to identify the size and number of bead loading regions best suited to yield the sequencing throughput for your DNA library sample. The throughput presented in Table 1 is typical for good 400 cycles sequencing Runs performed using good quality shotgun libraries and in Table 2 for amplicon libraries with fragments up to 700 bp. For example, column 4 of Table 1 shows the expected throughput of a full sequencing Run carried out with the longest run script available (400 cycles, generating reads up to 1 kbp in length), with all regions loaded. Region Size Regions per PTP Device Bases per Region (Mbp) Bases per Full PTP Device (Mbp) Reads per Region (x 10 3 ) Large Medium M/S Small Table 1: Gasket specification and throughput for shotgun libraries Region Size Regions per PTP Device Reads per Region (x 10 3 ) Large 2 >255 Medium 4 >100 M/S 8 >50 Small 16 >10 Table 2: Gasket specification and throughput for amplicon libraries 3 / 3
4 Use the following examples as guidance of gasket loading for your experiment: If the experiment aims to sequence 10 Mbp at 25-fold coverage, a total throughput of 250 Mbp is required. This can be obtained in a single large region of a PTP device. Sequencing 70 Mbp at a 30-fold coverage, requires just over 2 Gbp of total throughput and about 8 large regions, or 4 full sequencing Runs with the large region gasket. Resequencing 4 Mbp at 10-fold coverage yields a total 40 Mbp throughput and requires one single medium/small region. The Sequencing Reagents of the GS FLX Titanium Sequencing Kit XL+ must be thawed prior to use in the sequencing procedure described in this manual. Also, the bottle of Titanium Bead Buffer must be pre-chilled because it will be supplemented with Apyrase, a thermolabile enzyme, to make Bead Buffer 2. Before beginning an experiment, do the following: Library Sample Type on PTP Device: Libraries with different sequencing keys cannot be loaded in the same region of a PTP device. The PTP device can hold, in different regions, libraries with different sequencing keys. 1. Bring the Sequencing Reagents tray of the GS FLX Titanium Sequencing Kit XL+ out of its frozen storage. Keep the Sequencing Enzymes sleeve at -15 to -25ºC. 2. Remove the barrier bag from the Sequencing Reagents tray. Remove the tube of DTT from position 3 and place it with the Sequencing Enzymes sleeve at -15 to -25ºC. Allow the other reagents to thaw, using one of the following procedures, keeping the tray upright and protected from bright light. Immerse (do not allow the water level to reach the tube caps) in a room temperature water bath for 2 h. or Leave for 2.5 h at room temperature. or Leave at +2 to +8ºC for up to 60 h (e.g. weekend). 3. Before beginning, place the bottle of Titanium Bead Buffer on ice. 4. For room temperature thawing (first two bullets above), when the contents of the Sequencing Reagents tray are thawed, transfer the tray to +2 to +8ºC to keep the reagents chilled until the Run. Do not let the reagents remain at this temperature for more than 48 h. 5. Within 3 h of beginning the procedure, place the reagents of the Sequencing Enzymes sleeve to thaw on ice. Place the tube of DTT in a room temperature water bath to thaw, then leave at room temperature. Do not re-freeze the reagents. Two sequencing keys exist for libraries: TCAG for General, Paired End, cdna and Amplicon libraries GACT for Rapid, cdna Rapid and Paired End Rapid libraries Reagents and Titanium Bead Buffer 4/4
5 3 PROCEDURE 3.1 The Pre-Wash To login, select your user name in the Operator ID dropdown menu and click Sign In. The main window of the GS Sequencer application will open (Figure 3). Under normal conditions of continuous operation, the instrument is kept running. Therefore, the PTP device from the previous Run should still be in place in the instrument s PTP cartridge, within the camera door. If there is no PTP device in the cartridge, you must install a used but intact one (and a cartridge seal) before proceeding with the prewash. 1. Close the previous run by clicking OK in the Sequencing run complete window. 2. If required, log in as follows: Click on Operator in the Status area of the application window, Figure 2. Figure 3: The GS Sequencer application window after an Operator logs in Figure 2: The login window of the GS Sequencer application 5/5
6 3. Discard the spent reagents and clean the GS FLX+ Reagents Cassette, then perform the pre-wash: a. Open the exterior fluidics door and raise the sipper manifold completely (Figure 4A). b. Is it important to fill the Pre-wash Tubes to the top, as shown in Figure 5A, with Pre-wash Buffer, for the Pre-wash to proceed correctly. Slide out the spent Reagents Cassette (Figure 4B). A A B Figure 5: Filling the Pre-wash Tubes; loading before a pre-wash Figure 4: Removing the GS FLX+ Reagents Cassette (from the previous Run) c. Discard the spent bottles and tubes. d. e. f. Empty the GS FLX+ Waste Container and replace the cap. Dry all the surfaces of the cassette with a paper towel. Replace the Sipper Tubes as described in the GS FLX+ Instrument Owner s Manual. Change gloves to avoid contaminating other components. Do not wipe the filter nor the Luer connector of the manifold as this risks introducing fibers into the system, which could clog fluidics valves. To prepare the Pre-wash Cassette, place the GS FLX+ Pre-wash Tube Holder on top of the Reagents Cassette and insert five long, grey Pre-wash Tubes on the left side and ten short grey Pre-wash Tubes on the right side in the prewash tube holder. Fill all the tubes to the top with Pre-wash Buffer (as in Figure 5A) and slide the cassette in the instrument, lower the sipper manifold and close the fluidics door (Figure 5B). g. h. B 4. 6/6 If there isn t a PTP device or if the cartridge seal had leaked in the previous run (if there is evidence of liquid under the camera door), install a used but intact PTP device (with a used but intact cartridge seal) following instruction is Section
7 3.1.1 Launch the Pre-Wash 1. Return to the instrument computer. If the Instrument tab is not selected, select it now (Figure 6). 2. Click the Start button in the Global Action area. This opens the Run Wizard s first window: Choose a procedure (Figure 7). Figure 6: The Instrument tab of the GS Sequencer software Figure 7: The Run Wizard s first window (Choose a procedure), with the Pre-wash option selected 7 / 7
8 3. In the Run Wizard s first window, select the Pre-wash option, and click the Next button. This opens the Run Wizard s second window: Start Pre-wash (Figure 8). Monitor the instrument until the Status LED located above the camera door on the instrument is blinking green. If the instrument encounters any problems during the initiation of the Pre-wash, a message describing the issue will appear in the Status area of the GS Sequencer window. There is an Abort button available in the GS Sequencer software main window (Figure 6), which can be clicked if a problem occurs. PicoTiterPlate device preparation: Start the preparation of the PTP device (Section 3.2) as soon as the Pre-wash is safely ongoing. The Prewash will proceed to completion without any further user intervention (approximately 1 hour). Figure 8: The Run Wizard s second window (last Wizard window for Pre-wash): Start Prewash 4. Click the Start button in the Start Pre-wash window (from the Run Wizard) to start the Pre-wash operation. 8/8
9 3.2 PicoTiterPlate (PTP) Device Preparation Prepare Enzyme Bead Wash (EB Wash) and Bead Buffer 2 (BB2) 1. Add 1.2 ml of Titanium Supplement CB to the 200 ml bottle of pre-chilled Titanium Bead Buffer. Re-cap and mix gently by inversion. 2. Pour 20 ml of the buffer made in Step 1 in a 50 ml conical tube. Remove the tube of Apyrase from the ice. Leave the other components on ice until needed. 3. Add 6.8 µl of Apyrase to the 20 ml supplemented Titanium Bead Buffer in the conical tube. Re-cap and label the conical tube EB Wash. Mix gently by inversion, and place the conical tube on ice. 4. Add 30.6 µl of Apyrase to the bottle of supplemented Titanium Bead Buffer. Re-cap and label the bottle BB2. Mix gently by inversion, and place the bottle on ice. 5. Return the remainder of the Titanium Supplement CB and of the Apyrase on ice Prepare the PicoTiterPlate and Bead Deposition Devices 1. Retrieve the PTP device shipping tray. Lift the PTP device out of the shipping tray with a gloved hand and place it back into the tray. 2. Write down the PTP device ID to enter the 6-digit number in the second window of the Run Wizard (Section 3.3.4). 3. Completely submerge the PTP device in BB2 at room temperature until you are ready to assemble and spin the Bead Deposition Device (BDD) (Section ). Keep the rest of the BB2 on ice. 4. Wash the Bead Loading Gasket, by gently shaking the gasket and seal for 30 seconds in a Sparkleen solution (or nanopure water if Sparkleen is not available). Rinse thoroughly with nanopure water and let air dry on a paper towel. 5. Wash the appropriate BDD using a soft bristle brush and Sparkleen solution. Rinse thoroughly with nanopure water and let the device air dry on a paper towel Prepare the Beads The GS FLX+ System contains four kinds of microparticles (beads), as listed in Table 3. Each type of bead must undergo a specific preparation procedure, as described below. Bead Type DNA Beads Enzyme Beads PPiase Beads Packing Beads Description/Function DNA Beads carry the DNA library to be sequenced (sample). These sample DNA Beads have been prepared using the appropriate 454 Sequencing System procedures and kits. Before use, the sample DNA Beads will be spiked (Section ) with Control DNA Beads (included in the GS FLX Titanium Sequencing Kits), which serve as an internal control for the sequencing reaction. Control DNA Beads are also available in a GS FLX Titanium Control Bead Kit, which can be used to carry out a separate, test sequencing Run (see the Package Insert to the GS FLX Titanium Control Bead Kit). Enzyme Beads carry the immobilized enzyme components of the chemiluminescence system (sulfurylase and luciferase). PPiase Beads scavenge inorganic pyrophosphate (PPi) to reduce well-to-well crosstalk and interference during each nucleotide flow, as well as residual background noise after and between flows. Packing Beads stabilize and maintain all the immobilized components of the system within the wells of the PTP device, throughout the sequencing Run. Table 3: The four types of beads used in the GS FLX+ System 9 / 9
10 Prepare the Packing Beads Prepare the DNA Beads (Sample and Control) 1. Wash the Packing Beads three times, as follows: a. Add 1 ml of BB2 by centrifuge at 10,000 rpm (9300 x g RCF) for 5 minutes. Break up aggregates by vortexing, until a uniform suspension is achieved. b. Remove and discard the supernatant without disturbing the bead pellet c. Repeat Steps a and b two times. You may start preparing the DNA Beads during these three centrifugations. 2. After the third wash, add 550 µl of BB2 per tube and resuspend the beads and keep on ice. 1. Using Table 4, determine the number of sample DNA beads required for your gasket (column 2). 2. Calculate the volume of DNA library beads to be sequenced based on the number of beads needed and the concentration of the library, in beads/µl. 3. Vortex the DNA library beads and transfer the appropriate amount of beads into new tubes of the appropriate size (e.g. 2 ml tubes for large regions, 1.7 ml tubes for medium or M/S regions, and 0.2 ml tubes for small regions). Use a separate tube for each loading region. 4. Vortex the Control DNA Beads and add the appropriate volume of bead suspension to each DNA library bead tube (column 3). 5. Calculate the volume of supernatant to remove that will leave the volume indicated in column 4, for the bead loading gasket configuration you are using. This volume will vary depending on the volume of DNA library beads used, which in turn depends on the concentration of the DNA librarybeads. 6. Spin the beads for 1 minute to form a pellet (use a minifuge or a microcentrifuge). Rotate the tube 180 and spin again for 1 minute. If the total volume of bead suspension is equal to or less than the volume indicated in column 4, skip this centrifugation, as you have no volume to remove. 7. Set a pipettor to the volume calculated in Step 5, above. Being careful not to disturb the bead pellet, carefully and slowly remove the appropriate amount of supernatant and discard it. Loading Region Size Number of DNA Library Beads to Load per Region Volume of Control DNA Beads (µl) Target Final Volume, after Centrifugation (µl) Large 2,000,000 (x 2) 20 (x 2) 50 (x 2) Medium 790,000 (x 4) 10 (x 4) 30 (x 4) Medium/Small 340,000 (x 8) 6 (x 8) 15 (x 8) Small 125,000 (x 16) 3 (x 16) 10 (x 16) Table 4: Preparation of the DNA Beads, for each bead loading gasket configuration 10 / 10
11 8. Separately, prepare the DNA Bead Incubation Mix (DBIM) in a clean 15 ml tube (Table 5). Vortex to ensure that the Polymerase Cofactor and DNA Polymerase are thoroughly mixed. Bulk preparation of DBIM is fine, even for gaskets with multiple loading regions. The proper amount will be pipetted into individual tubes of DNA beads in the next step. Loading Region Size BB2 (µl) Polymerase Cofactor (µl) DNA Polymerase (µl) Total Volume (µl) All sizes These two types of beads can be washed in parallel, in separate tubes. However, make sure to change pipette tips to avoid contaminating them with one another. Table 5: Preparation of the DNA Bead Incubation Mix 9. Add the DBIM to each tube of DNA beads, per Table 6. Vortex well. If the volume of DNA beads in your tubes is less than the volume indicated in the second column, add extra DBIM so each tube contains the Total Volume indicated in the last column. The leftover DBIM can be discarded. Loading Region Size DNA Beads (µl) DBIM (µl) Total Volume (µl) Large 50 (x2) 950 (x 2) 1000 (x 2) Medium 30 (x4) 320 (x 4) 350 (x 4) Medium/Small 15 (x8) 140 (x8) 155 (x 8) Small 10 (x 16) 50 (x 16) 60 (x 16) Prepare the Enzyme and PPiase Beads (Bead Layers 1, 3 & 4) 1. Vortex the Enzyme Beads and the PPiase Beads and place them in a Magnetic Particle Concentrator (MPC) for at least 2 minutes for the beads to pellet. Invert the MPC several times and wait at least 2 minutes again. Carefully remove the supernatants and then remove the tubes from the MPC. 2. Wash the Enzyme Beads three times with 1 ml of EB Wash for each tube of Enzyme Beads. Vortex and wash the beads using the MPC, as above. 3. After the third wash, add 1 ml of EB Wash to each tube of Enzyme Beads. Vortex to resuspend the pellets. Keep on ice. 4. Wash the PPiase Beads three times with 1 ml of BB2. Vortex and wash the beads using the MPC, as above. 5. After the third wash, add 500 µl of BB2 to the tube of PPiase Beads. Vortex to resuspend the pellets. Keep on ice. 6. Prepare the beads for layers 1, 3, and 4 in three separate appropriately labeled tubes of an appropriate size (e.g. 15 ml). This is done by diluting the Enzyme Beads (layer 1: Enzyme Beads pre-layer; and layer 3: Enzyme Beads post-layer) and the PPiase Beads (layer 4) in EB Wash or BB2, as listed in Table 7. Vortex the washed Enzyme and PPiase Beads to get a uniform suspension before transferring. Bead Layer EB Wash or BB2 (µl) Enzyme Beads (µl) PPiase Beads (µl) Total Volume (µl) (EB Wash) (EB Wash) (BB2) Table 6: Dilution of DNA Beads in DNA Bead Incubation Mix 10. Incubate the samples on the lab rotator at room temperature for 15 minutes. It is convenient to prepare the beads of layers 1, 3, and 4 during this 15 min of incubation. Do not exceed 50 min for this incubation. Table 7: Dilution of the Enzyme and PPiase Beads for Bead Layers 1, 3, and 4 11 / 11
12 Combine the DNA and Packing Beads (Bead Layer 2) After the DNA beads incubation in DBIM is complete, return to the washed Packing Beads and the leftover BB2. Vortex and transfer the appropriate volumes of washed Packing Beads and of BB2 to the tubes containing the DNA beads, as listed in Table 8. Assemble the BDD with the PTP Device and Bead Loading Gasket 1. Using a gloved hand, remove the PTP device from the BB2 bath, handling it by the edges only, and pour off excess BB2. 2. Wipe the back of the PTP device with a Kimwipe. However, be careful NOT to allow the side with the wells to dry. Loading Region Size DNA Beads (µl) Packing Beads (µl) BB 2 (µl) Total Volume (µl) 3. Place the PTP device onto the BDD base (Figure 9A), aligning the notched corners of the PTP device and the BDD base. Large 1000 (x 2) 265 (x 2) 435 (x 2) 1700 (x 2) 4. Medium 350 (x 4) 100 (x 4) 210 (x 4) 660 (x 4) M/S 155 (x 8) 45 (x 8) 125 (x 8) 325 (x 8) Secure the washed and dried bead loading gasket to the BDD base by laying it on top of the PTP device and placing its molded ridge into the groove on the BDD base (Figure 9B). Align the notched corners of the bead loading gasket and the BDD base, as shown in the Figure. Small 60 (x 16) 15 (x 16) 35 (x 16) 110 (x 16) 16 Small Regions Bead Loading Gasket: Table 8: Preparation of Bead Layer 2 2. The region separators of this gasket sometimes stick together. If this happens, gently separate them manually before laying the gasket on top of the PTP device, in the BDD. Vortex the DNA and Packing Beads mixes and place them on the lab rotator for at least 5 min at room temperature. The unused Packing Beads can be discarded. After installing this gasket in the BDD, gently spread it lengthwise to ensure that the lanes are straight and uniform. 12 / Carefully place the BDD top over the assembled BDD base/ptp device/gasket (Figure 9C) so the port holes and air vents line up with the loading regions defined by the gasket, underneath. Align the dowels on the BDD base so they slide into the holes in the BDD top and the BDD top is sitting flat across the PTP device. 6. Press down on the top of the BDD, and rotate the two latches from the BDD base into the grooves in the BDD top to firmly secure the assembly (Figure 9D). When you hear a click, the latches should be firmly seated in the grooves, providing the correct amount of pressure to maintain a liquid-tight seal.
13 A B Deposit the Four Layers of Buffer/Beads on the PTP Device Deposit the beads onto the PTP device by injecting the suspensions through the port holes of the assembled Bead Deposition Device (BDD), and then using centrifugal sedimentation to settle the beads to the bottom of the PicoTiterPlate wells. The PTP device is wetted first with BB2, and this process is then repeated for each of four layers: C D Wet the PTP device with Bead Buffer 2 Bead Layer 1: Enzyme Beads pre-layer Bead Layer 2: DNA and Packing Beads layer Bead Layer 3: Enzyme Beads post-layer Bead Layer 4: PPiase Bead layer Pipette tips: Use the correct pipette tips to load the beads: 2 ml tip for Large regions, 1 ml tip for Medium and M/S regions, and 200 µl tip for Small regions. Time between centrifugations: Minimize the time interval between loading the beads and starting the centrifugation. Figure 9: Assembly of the Bead Deposition Device Bead distribution: Load the beads promptly for an even distribution in the PTP device. Air bubbles: Avoid injecting air into the BDD. Bead delivery: Use a single, even injection to fill each loading region of the BDD. Loading region fill: Fill the BDD completely but do not overflow the loading regions. Discard any excess bead mix. 13 / 13
14 Wet the PTP device: Bead Buffer 2 1. Using a pipette and a tip of the proper size, draw the appropriate amount of BB2 for the loading region size you are using per Table 9, and add the buffer through the loading ports. Loading Region Size Large 1860 (x 2) Medium 660 (x 4) Medium/Small 325 (x 8) Small 110 (x 16) Volume to Load (µl) Table 9: Volume of BB2 to load per region for each bead loading gasket configuration Loading Region Size Large 1860 (x 2) Medium 660 (x 4) Medium/Small 325 (x 8) Small 110 (x 16) Volume to Load (µl) Table 10: Volume of Bead Layer 1 to load per region for each bead loading gasket configuration 4. Promptly load the bead suspension onto the first region of the PTP device, through the port hole on the BDD top (see Figure 10). Make sure to use a single, smooth dispensing action to ensure even distribution of the beads over the entire region of the PTP device. 2. Place both the assembled BDD and the BDD counterweight into centrifuge swinging baskets as appropriate for your centrifuge rotor, and place them onto the rotor opposite each other. 3. Centrifuge the loaded PTP device in the BDD for 5 minutes at 1620 x g (2640 RPM for the Beckman Coulter X-12 or X-15 centrifuges) Deposit Bead Layer 1: the Enzyme Beads Pre-Layer 1. Pipet out BB2 from the loading ports. 2. Vortex the bead suspension for Bead Layer 1 for 5 seconds to obtain a homogeneous suspension. 3. Using a pipette and tip of the proper size, draw the appropriate amount of bead suspension for the loading region size you are using, per Table 10. Figure 10: Filling the loading regions of the assembled Bead Deposition Device 5. Repeat Steps 1 to 4 for all loading regions of the PTP device. 6. Place both the assembled BDD and the BDD counterweight in centrifuge swinging baskets as appropriate for your centrifuge rotor, and place them on the rotor opposite each other. 7. Centrifuge the loaded PTP device in the BDD for 5 minutes at 1620 x g (2640 RPM for the Beckman Coulter X-12 or X-15 centrifuges). 14 / 14
15 Step synchronization: Perform the steps of Section during this 5 minutes centrifugation Step synchronization: Perform the Steps of Section during this 10 minutes of centrifugation. Deposit Bead Layer 2: the DNA and Packing Beads Deposit Bead Layer 3: the Enzyme Beads Post-Layer 1. Remove the BDD from the centrifuge. 1. Change gloves after loading the Reagents Cassette into the instrument. 2. With a pipettor, gently remove as much of the supernatants as possible through the port holes on the BDD top. 2. Remove the BDD from the centrifuge. 3. With a pipettor, gently draw out all the supernatant from the centrifuged Bead Layer 2, through the port holes on the BDD top. It is useful to save this Bead Layer 2 supernatant, for troubleshooting purposes in case the sequencing Run does not produce the expected number of reads. 4. Vortex the bead suspension for Bead Layer 3 for 5 seconds to obtain a uniform suspension. 5. Using a pipette and tip of the proper size, draw the appropriate amount of bead suspension for the loading regions you are using, per Table Remove the tubes of bead suspension for Bead Layer 2 from the rotator. 4. Briefly spin the bead suspension for Bead Layer 2 to collect material from the cap. Pipette up and down to mix. 5. Using a pipette and tip of the proper size, draw the appropriate amount bead suspension for the loading region size you are using, per Table 11. Loading Region Size Volume to Load (µl) Large 1700 (x 2) Medium 660 (x 4) Medium/Small Small Loading Region Size Volume to Load (µl) 325 (x 8) Large 1860 (x 2) 110 (x 16) Medium 660 (x 4) Medium/Small 325 (x 8) Small 110 (x 16) Table 11: Volume of Bead Layer 2 to load per region for each bead loading gasket configuration 6. Promptly load them onto the first region of the PTP device, through the port hole on the BDD top (see Figure 10). 7. Repeat Steps 4 to 6 for all loading regions of the PTP device. 8. Centrifuge the loaded PTP device in the BDD for 10 minutes at 1620 x g RCF (2640 rpm for the X-12 or the X-15 centrifuges). Table 12: Volume of Bead Layer 3 to load per region for each bead loading gasket configuration 15 / 15
16 For the third and fourth Bead Layers, these volumes may be more than needed to fill the loading regions, due to remaining fluid volume leftover on the PTP device. This is not a problem. 6. Promptly load the bead suspension onto the first region of the PTP device, through the port hole on the BDD top (see Figure 10). 7. Repeat Steps 4 to 6 for all loading regions of the PTP device. 8. Centrifuge the loaded PTP device in the BDD for 5 minutes at 1620 x g RCF (2640 rpm for the Beckman Coulter X-12 or X-15 centrifuges). Remove the BDD from the centrifuge 2. With a pipettor, gently draw out and discard all the supernatant from the centrifuged Bead Layer 3, through the port holes on the BDD top. 3. Vortex the bead suspension for Bead Layer 4 for 5 seconds to obtain a uniform suspension. 4. Using a pipette and tip of the proper size, draw the appropriate amount of bead suspension for the loading regions you are using, per Table 13. Large 1860 (x 2) Medium 660 (x 4) Medium/Small 325 (x 8) Small 110 (x 16) 5. Promptly load the bead suspension onto the first region of the PTP device, through the port hole on the BDD top (see Figure 10). 6. Repeat Steps 3 to 5 for all loading regions of the PTP device. 7. Centrifuge the loaded PTP device in the BDD for 5 minutes at 1620 x g RCF (2640 rpm for the Beckman Coulter X-12 or X-15 centrifuges). Step synchronization: By the time you reach this point in the procedure, you should have already performed the steps of Sections through If you have not, complete these steps now. If you have, proceed with Section You will collect the fully prepared PTP device from the centrifuge in Step 4 of that Section. Deposit Bead Layer 4: the PPiase Beads 1. Volume to Load (µl) Table 13: Volume of Bead Layer 4 to load per region for each bead loading gasket configuration Step synchronization: Perform the steps of Sections and during this 5 minutes centrifugation Loading Region Size 16 / 16
17 3.3 The Sequencing Run Remove the Pre-Wash Cassette and Clean the Fluidics Area Deck The Pre-wash Tubes should be discarded after every run. Step synchronization: Perform the procedures described in this Section during the 5 minutes centrifugation of the Bead Layer On the instrument computer, a Pre-wash run complete message will be displayed in the Status area of the GS Sequencer software main window. Click OK to continue. 2. Open the exterior fluidics door and raise the sipper manifold completely (see Figure 11A). 3. Slide out the Pre-wash Reagents Cassette (Figure 11B). A 5. Wipe all the surfaces of the Reagents Cassette with a paper towel. 6. Wipe the fluidics area deck (inside the instrument) with 50% ethanol and a paper towel, being careful to not touch the Sipper Tubes. Allow the deck to air dry completely. Sipper Tubes contamination risk: Never touch the Sipper Tubes unless you are replacing them. If contamination of any of the lines is suspected, replace them by following the instructions provided in the GS FLX+ Instrument Owner s Manual. B Step synchronization: Return to the bead deposition procedure for the DNA and Packing Beads layer (Bead Layer 2, Section ). Figure 11: Removing the Pre-wash cassette from the instrument 4. Remove the grey Pre-wash Tubes and the GS FLX+ Pre-wash Tube Holder from the Reagents Cassette. Empty the tubes in a sink and discard them. 17 / 17
18 3.3.2 Prepare and Load the Sequencing Reagents Cassette Step synchronization: Perform the procedures described in this Section during the 10 minutes centrifugation of the DNA and Packing Beads layer (Bead Layer 2). Chilled reagents: The concentrated reagents of the Sequencing Reagents tray MUST be thawed but kept refrigerated (+2 to +8 C). Make sure that the DTT is completely resuspended before supplementing. 1. Retrieve the five bottles of Titanium Buffer CB stored at room temperature. 2. Add 13.2 ml of Titanium Supplement CB and 2 ml of DTT to each bottle of Titanium Buffer CB. Re-cap and gently invert the bottles to mix. 3. Retrieve the Sequencing Reagents tray from +2 to +8 C. 4. Dilute 5 µl of the PPiase reagent in 45 µl Inhibitor TW reagent (position 9) in a 1.7 ml tube, and vortex. 5. Supplement the two reagents from the Sequencing Reagents tray with their appropriate additives, as listed below; then, change gloves. a. Add 32 µl of diluted PPiase (from Step 4) to the tube of Inhibitor TW (tray position 9, pink sticker). b. Add 675 µl of Apyrase to the tube of Apyrase Reagent Buffer (tray position 11, yellow sticker). 6. Make sure that all the caps are secure and invert the tray 20 times to uniformly mix the contents of all tubes. Make sure that there are no undissolved particulates in any of the reagents (pay particular attention to the tube of Substrate reagent). 7. Slide the empty Reagents Cassette at least half-way in the instrument, with the Sequencing Reagents tray platform placed on the right (Figure 12). Make sure that the tubes are fully pushed down, with the caps resting on the tray. Figure 12: The empty Reagents Cassette in the GS FLX+ Instrument 8. Loosen the caps of all the bottles and tubes. 9. Place the five bottles of supplemented Titanium Buffer CB XL+ on the left side of the cassette (Figure 13A). 10. Place the Sequencing Reagents tray on its platform on the right side of the cassette, with the tube in position 11 resting on top of the retainer block (Figure 13B). A B Figure 13: The Sequencing Reagents tray in the proper orientation 18 / 18
19 11. Remove the caps from the bottles and tubes, beginning by the ones in the back, avoiding handling caps over opened containers and touching the Sipper Tubes. Sensitive camera face: Always be extremely careful when handling or working near the camera face. Never touch the camera face with anything other than Zeiss moistened cleaning tissue or Lens paper from Thorlabs. 12. Slide the full Reagents Cassette completely in the instrument (Figure 14A), making sure that the tube of Apyrase (position 11) is properly engaged in the chiller. Lower the sipper carefully, and close the exterior fluidics door (Figure 14B). Change gloves before proceeding with depositing Bead Layer 3. A B 1. Click the Unlock button near the lower-left corner of the Instrument Tab (Figure 15). Figure 14: Loading the Reagents Cassette into the instrument Figure 15: The Unlock button used to unlock the camera door Step synchronization: Return to the bead deposition procedure for the Enzyme Beads post-layer (Bead Layer 3; Section ) Open the camera door by pulling gently on the handle as shown in Figure 16. Clean the PicoTiterPlate Cartridge and the Camera Faceplate Step synchronization: Perform the procedures described in this Section and in Section during the 5 minutes centrifugation of the Enzyme Beads post-layer (Section , Step 8). Figure 16: Opening the camera door 19 / 19
20 3. Remove the spent PTP device from the PTP cartridge by first pressing the PTP frame spring latch (Figure 17A) to lift the frame from the cartridge, and then sliding out the used PTP device (Figure 17B). A B Load and Set the Run Script and Other Run Parameters (without LIMS) This Section describes how to manually set up and launch a sequencing Run. However, if the GS FLX+ Instrument is connected to a Laboratory Information Management System (LIMS), the Run Wizard will seek the information that describes the Run from your LIMS after you enter the PTP device ID. Ensure there is enough free data space on the instrument for the run. Go to the Data tab and click on the instrument icon at the top of the list of Runs to view the information about the free space available. The total size of the raw images for a 2 regions, 400 cycle GS FLX Titanium Sequencing Kit XL+ Run is about 35 GB. Figure 17: Removing the used PicoTiterPlate device 4. Close the PTP frame and, using a pair of plastic forceps provided in the accessory set, remove the used PTP cartridge seal from the PTP cartridge (Figure 18A). 5. Wet a Kimwipe with 50% ethanol and wipe the surface of the cartridge to remove any bead and reagent residue (Figure 18B). Allow the cartridge to air dry completely. A 1. B Figure 19: Upper portion of the GS Sequencer software main window, with the Instrument tab active Figure 18: Removing the PTP cartridge seal and wiping the PTP cartridge On the instrument computer, the GS Sequencer main window is displayed, with the Instrument Tab active (Figure 19). 20 / 20
21 2. Click the Start button in the Global Action area. This opens the Run Wizard s first window: Choose a procedure (Figure 20). 3. In the Run Wizard s first window, select the Sequencing Run option, and click the Next button. This opens the Run Wizard s second window: Enter IDs and bar codes Enter IDs and bar codes (Figure 21). Figure 20: The Run Wizard s first window: Choose a procedure, with Sequencing Run option selected Figure 21: The Run Wizard s second window: Enter IDs and bar codes 21 / 21
22 4. Enter the bar code of the PicoTiterPlate device to be used in this Run. You may also enter the product IDs or bar codes of other materials associated with this sequencing Run (e.g. Library Prep, empcr, and Sequencing Kits); you will be asked for this information if you call Roche Customer Support for help if you encounter any difficulties with your Run. When you have entered all the information, click Next. This opens the Run Wizard s third window: Enter Run name and Run Group (Figure 22). 5. Enter a specific, unique name for this Run. Then find and select your Run Group in the Run group list. Click Next. This opens the Run Wizard s fourth window: Choose sequencing kit (Figure 23). Figure 23: The Run Wizard s fourth window: Choose sequencing kit. Your selection will be highlighted. Figure 22: The Run Wizard s third window: Enter Run name and Run Group 22 / 22
23 6. Select XL+ and click Next. This opens the Run Wizard s fifth window: Choose PicoTiterPlate type (Figure 24). Flow pattern A is a cyclic TACG flow pattern that generates results similar to those obtained with a 400 cycle run in version 2.6 software. Flow pattern B is an acyclic flow pattern that is expected to increase read length after all signal processing filters have been applied. Flow pattern B is recommended for most genomic shotgun runs, and for long amplicon processing Sample Type Recommended Flow Pattern Expected Average Read Length Genomic shotgun Flow pattern B (Acyclic) Longer than both XLR70 and XL+ with flow pattern A Long Amplicon (>550 bp) Flow pattern B (Acyclic) Longer than XLR70 Transcriptomic (cdna) Flow pattern A (Cyclic) Flow pattern B (Acyclic) XLR70 read lengths Figure 24: The Run Wizard s fifth window: Choose PicoTiterPlate type 7. Select the bead loading gasket to be used in this Run, and click Next. This opens the Run Wizard s sixth window: Choose the flow pattern (Figure 25). 23 / 23
24 8. Select the flow pattern and click Next. a. This opens the Run Wizard s seventh window: Choose Run processing type (Figure 26). Figure 25: The Run Wizard s sixth window: Choose the flow pattern Figure 26: The Run Wizard s seventh window: Choose Run Processing type 24 / 24
25 9. Select the processing scheme appropriate for this Run, and click Next. Two data processing schemes are available on the GS FLX+ Instrument when using a GS FLX Titanium Sequencing Kit XL+ (see the 454 Sequencing System Software Manual for a complete description of the GS FLX+ System s data processing and analysis processes, including a description of when to use each of these options): Image processing only: The instrument will only capture images during the Run, and process them into raw wells data files; this will start during Run-time and may complete up to 3 hours or more after completion of the fluidics part of the sequencing Run. The read flowgrams and basecalls with per-base quality scores must then be produced later. No processing: The instrument will only capture and store raw images for this Run. All steps of data processing must then be carried out separately. Of the processing schemes shown on the Choose Run processing type window (Figure 26), only two are available during a sequencing run. All others are greyed and are not supported on the instrument. For additional information on the availability of various processing schemes, see Section of the 454 Sequencing System Software Manual. For additional technical details about the underlying data processing pipelines, see Section 1 of the 454 Sequencing System Software Manual. This opens the Run Wizard s eighth window: Request data backup (Figure 27). Figure 27: The Run Wizard s eighth window: Request data backup 25 / 25
26 10. Select the Backup Run upon completion checkbox, and click Next. With this checkbox selected, the data from the Run (including any data processing data) will be automatically backed up at the end of the Run (to a storage location pre-determined by an Administrator. See Appendix, Section 4). This opens the Run Wizard s ninth window: Run comments (Figure 28). 11. Enter any comments about the Run, and click Next. This opens the tenth window: Confirmations (Figure 29) Figure 29: The Run Wizard s tenth window: Confirmations Figure 28: The Run Wizard s ninth window: Run comments 26 / 26
27 12. Empty the waste container and check the box, then click Next. This opens the eleventh and last window: Insert new PicoPiterPlate device (Figure 30) Insert the PicoTiterPlate Device and Launch the Sequencing Run 1. Wet a clean Kimwipe with 10% Tween-20 and wipe the surface of the cartridge. 2. Install the cartridge seal as described below: a. b. A Make sure that the square ridge on the seal is facing up (see Figure 31A), and drop the seal in the cartridge groove (Figure 31B). If necessary, gently tap the seal into place with a gloved hand. DO NOT wipe the seal with anything, not even with a Kimwipe, as this could damage or stretch the seal. B Figure 31: Orientation and placement of the cartridge seal Figure 30: The Run Wizard s eleventh, and last window: Insert new PicoTiterPlate device Step synchronization: Return to the bead deposition procedure for the PPiase Beads layer (Bead Layer 4, Section ). 27 / 27
28 3. Press the PTP cartridge spring latch (Figure 32A) to lift the PTP frame from the cartridge. 4. When centrifugation of the Bead Layer 4 is complete (from Section , Step 7), remove the BDD from the centrifuge, and discard the BDD port seals that cover the loading ports and air vents. 5. With a pipettor, gently draw out and discard all the supernatant from the centrifuged Bead Layer 4, through the port holes in the BDD top. 6. Remove the PTP device from the BDD, as follows: a. Rotate down the latches of the BDD to unfasten them. b. Carefully remove the BDD top. c. Gently lift off and discard the bead loading gasket. d. Remove the PTP device, being careful to handle it only by the edges. 7. Slide the PTP device into the PTP cartridge frame, face down. Make sure that the PTP device notch is on the lower right hand corner, matching the notch in the frame (Figure 32B). 8. Close the PTP frame, making sure it is properly secured by the latch (Figure 32C). 9. Wipe the back of the PTP device with a Kimwipe. Use a downward motion, toward the camera faceplate, to avoid sliding the PTP device within the frame, or out of it. A B C D 10. Use a new Zeiss pre-moistened cleaning tissue to gently wipe the camera faceplate. 11. Allow the camera faceplate to air dry completely. 12. Close the camera door (Figure 32D). Figure 32: Loading a PTP device into the PTP frame, in the cartridge 13. Click the Start button in the Run Wizard s Insert new PicoTiterPlate device window (Figure 30) to start the sequencing Run. The Operator can monitor the progress of a sequencing Run by viewing the Instrument status and the data images as they are being captured by the camera: Thumbnail images will appear under the progress bar in the Instrument tab during the Run (Figure 33). If the Start button is still grayed out, check for x in a red circle icons in the Run Wizard window, and address any problems that may be reported by the sensors. 28 / 28
29 Monitor the instrument until the sequencing Run is under way and the Status LED located above the camera door on the instrument is blinking green (also shown at the upper-left corner of the GS Sequencer window). If the instrument encounters any problems during the initiation of the Run, a message describing the issue will appear in the Status area of the GS Sequencer window, and user intervention will be required. In most problem situations, the software will offer to Abort or Proceed with the Run. If you are certain that the warning is unwarranted then you can decide to proceed. In a few special cases, however, you will not be offered the possibility to proceed. If this happens, you must restart the software, as follows: Close the GS Sequencer application Double-click the systemstart icon, located on the desktop Re-launch the GS Sequencer, and set up your Run again. There is an Abort button available in the Global Action area of the GS Sequencer main window (Figure 33), which can be clicked if a problem occurs during the Run. The Abort Run dialog box will ask you to verify that you want to stop the Run; if you confirm, the Run will be immediately terminated. There is no procedure for pausing and resuming a Run. If a Run is aborted, follow the abort with a Pre-wash before proceeding with another sequencing Run. Figure 33: The Instrument tab of the GS Sequencer application main window during a sequencing Run 14. When the sequencing Run is complete, a message will appear in the Status area of the GS Sequencer application window. 29 / 29
30 4 APPENDIX: NETWORK S ETUP FOR SEQUENCING DATA BACKUP Data transfer is usually setup at the time of instrument installation. It is very highly recommended to setup a network data backup mechanism to store sequencing Run data, because the data sets created by 400 cycle XL+ Runs are large, and the storage capacity on the instrument is limited. In addition, data processing on the instrument would take a substantial amount of time (up to a week or more). To enable backup on a per Run basis, the instrument control software will execute a shell script at the completion of the Run. This script (/usr/local/rig/bin/backupscript.sh) is intended to be customer-configurable to execute any on-instrument process necessary to initiate the backup mechanism. Sequencing Kit Flow Pattern Run Length Raw Images Only Raw Images Plus Image Processing Raw Images Plus Full Processing* XL+ Flow Pattern A (Cyclic) 400 cycles 1600 nt flows ~35 GB 38 GB 60 GB XL+ Flow Pattern B (Acyclic) 1779 nt flows ~ 38 GB 45 GB 68 GB Table 14: Approximate amount of data generated by the XL+ sequencing kit. These numbers assume that the 2 regions gasket is used. nt: nucleotide. The script will have one argument passed to it: the fully qualified path of the Run directory. * For off instrument as Full Processing is not supported on the instrument. The only other requirement of the script is that its return value reflect the outcome of the backup procedure. This way, the instrument control GUI (the GS Sequencer application; the Data tab) can indicate to the user whether or not a particular Run s backup procedure was successful. Thus, the script should be modified such that the variable RET_VAL is set to 0 if the backup was successful and to 1 if it was not. Note that if Run backup was selected during setup, and the backup either did not take place or failed, the user will not be allowed to delete the Run. This behavior is described in the Section on the Data tab of the GS Sequencer application, in the 454 Sequencing System Software Manual. The Run data consists of image data, log files, and possibly image and signal processing data, if the Image Processing Only data processing scheme was chosen during Run setup. Table 14 shows the approximate size of the data sets created during a sequencing Run for each data processing option (assuming the largest bead loading regions available on the PTP device). Note that the space required to perform these runs is smaller than it would have with previous versions of the 454 Sequencing System software. Version 2.6 of the software introduced image compression that allows a 40% reduction in space required to store the raw data. 30 / 30
31
32 Table of Contents 1 Workflow Before You Begin What You Should Have Before Starting Sample Required GS FLX+ System Equipment and Reagents Kits Choosing the Size and Number of PTP Regions for Your Sample Reagents and Titanium Bead Buffer Procedure The Pre-Wash Launch the Pre-Wash PicoTiterPlate (PTP) Device Preparation Prepare Enzyme Bead Wash (EB Wash) and Bead Buffer 2 (BB2) Prepare the PicoTiterPlate and Bead Deposition Devices Prepare the Beads Prepare the Packing Beads Prepare the DNA Beads (Sample and Control) Prepare the Enzyme and PPiase Beads (Bead Layers 1, 3 & 4) Combine the DNA and Packing Beads (Bead Layer 2) Assemble the BDD with the PTP Device and Bead Loading Gasket Deposit the Four Layers of Buffer/Beads on the PTP Device Wet the PTP device: Bead Buffer Deposit Bead Layer 1: the Enzyme Beads Pre-Layer Deposit Bead Layer 2: the DNA and Packing Beads Deposit Bead Layer 3: the Enzyme Beads Post-Layer Deposit Bead Layer 4: the PPiase Beads The Sequencing Run Remove the Pre-Wash Cassette and Clean the Fluidics Area Deck Prepare and Load the Sequencing Reagents Cassette Clean the PicoTiterPlate Cartridge and the Camera Faceplate Load and Set the Run Script and Other Run Parameters (without LIMS) Insert the PicoTiterPlate Device and Launch the Sequencing Run Appendix: Network Setup for Sequencing Data Backup...30 Published by 454 Life Sciences Corp. A Roche Company Branford, CT USA Life Sciences Corp. All rights reserved. Notice to Purchaser For patent license limitations for individual products please refer to: For life science research only. Not for use in diagnostic procedures. Trademarks 454, 454 LIFE SCIENCES, 454 SEQUENCING, GS FLX, GS FLX TITANIUM, GS JUNIOR, EMPCR, PICOTITERPLATE, and PTP are trademarks of Roche. All other product names and trademarks are the property of their respective owners. (3) 0613
Sequencing Method Manual for the GS FLX+ Instrument
 Sequencing Method Manual for the GS FLX+ Instrument GS FLX+ Series XLR70 Kit May 2011 For life science research only. Not for use in diagnostic procedures. Instrument / Kit GS Junior / Junior GS FLX+ /
Sequencing Method Manual for the GS FLX+ Instrument GS FLX+ Series XLR70 Kit May 2011 For life science research only. Not for use in diagnostic procedures. Instrument / Kit GS Junior / Junior GS FLX+ /
GS FLX Titanium Sequencing Method Manual. October 2008
 GS FLX Titanium Sequencing Method Manual October 2008 798_Seq-MethMan_Titel-RS.indd 1 13.08.2008 12:07:28 Uhr Table of Contents Preface About this Manual...5 Safety...7 Assumptions...8 Sample ready...8
GS FLX Titanium Sequencing Method Manual October 2008 798_Seq-MethMan_Titel-RS.indd 1 13.08.2008 12:07:28 Uhr Table of Contents Preface About this Manual...5 Safety...7 Assumptions...8 Sample ready...8
empcr Amplification Method Manual Lib L LV
 empcr Amplification Method Manual Lib L LV GS FLX+ Series XL+ May 2011 For life science research only. Not for use in diagnostic procedures. Instrument / Kit GS Junior / Junior GS FLX+ / XL+ GS FLX+ /
empcr Amplification Method Manual Lib L LV GS FLX+ Series XL+ May 2011 For life science research only. Not for use in diagnostic procedures. Instrument / Kit GS Junior / Junior GS FLX+ / XL+ GS FLX+ /
DNA extraction Protocol for Agencourt Genfind v2 Blood and Serum Genomic DNA Isolation Kit
 DNA extraction Protocol for Agencourt Genfind v2 Blood and Serum Genomic DNA Isolation Kit Introduction The Agencourt Genfind v2 Blood & Serum DNA Isolation Kit utilizes Agencourt s patented SPRI paramagnetic
DNA extraction Protocol for Agencourt Genfind v2 Blood and Serum Genomic DNA Isolation Kit Introduction The Agencourt Genfind v2 Blood & Serum DNA Isolation Kit utilizes Agencourt s patented SPRI paramagnetic
AGENCOURT GENFIND Blood & Serum Genomic DNA Isolation Kit
 Blood & Serum Genomic DNA Isolation Kit Page 1 of 9 Please refer to http://www.agencourt.com/technical for updated protocols and refer to MSDS instructions when handling or shipping any chemical hazards.
Blood & Serum Genomic DNA Isolation Kit Page 1 of 9 Please refer to http://www.agencourt.com/technical for updated protocols and refer to MSDS instructions when handling or shipping any chemical hazards.
Procedure & Checklist >20 kb Template Preparation Using BluePippin Size-Selection System (15-20 kb Cutoff) for Sequel Systems
 Procedure & Checklist >20 kb Template Preparation Using BluePippin Size-Selection System (15-20 kb Cutoff) for Sequel Systems Before You Begin To perform this procedure, you must have the PacBio Template
Procedure & Checklist >20 kb Template Preparation Using BluePippin Size-Selection System (15-20 kb Cutoff) for Sequel Systems Before You Begin To perform this procedure, you must have the PacBio Template
Procedure & Checklist - 20 kb Template Preparation Using BluePippin Size-Selection System
 Procedure & Checklist - 20 kb Template Preparation Using BluePippin Size-Selection System Before You Begin To perform this procedure, you must have the PacBio DNA Template Prep Kit 2.0 (3 kb to 10 kb)
Procedure & Checklist - 20 kb Template Preparation Using BluePippin Size-Selection System Before You Begin To perform this procedure, you must have the PacBio DNA Template Prep Kit 2.0 (3 kb to 10 kb)
SOLiD EZ Bead Emulsifier
 QUICK REFERENCE SOLiD EZ Bead Emulsifier Pub. Part no. 4441487 Rev. E Rev. Date October 2011 Note: For safety guidelines, refer to the Safety section in the Applied Biosystems SOLiD EZ Bead Emulsifier
QUICK REFERENCE SOLiD EZ Bead Emulsifier Pub. Part no. 4441487 Rev. E Rev. Date October 2011 Note: For safety guidelines, refer to the Safety section in the Applied Biosystems SOLiD EZ Bead Emulsifier
Procedure and Checklist - 20 kb Template Preparation Using BluePippin Size-Selection System
 Procedure and Checklist - 20 kb Template Preparation Using BluePippin Size-Selection System Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit and have reviewed the
Procedure and Checklist - 20 kb Template Preparation Using BluePippin Size-Selection System Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit and have reviewed the
Procedure & Checklist - 1 kb Template Preparation and Sequencing
 Procedure & Checklist - 1 kb Template Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit. Fragment and Concentrate DNA Important: The distribution
Procedure & Checklist - 1 kb Template Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit. Fragment and Concentrate DNA Important: The distribution
Procedure & Checklist - 2 kb Template Preparation and Sequencing
 Procedure & Checklist - 2 kb Template Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio DNA Template Prep Kit (verify you have the correct kit for your insert
Procedure & Checklist - 2 kb Template Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio DNA Template Prep Kit (verify you have the correct kit for your insert
Select-a-Size DNA Clean & Concentrator MagBead Kit Catalog No. D4084 & D4085
 INSTRUCTION MANUAL Select-a-Size DNA Clean & Concentrator MagBead Kit Catalog No. D4084 & D4085 Highlights Tunable: Size selection can be tuned from 100 bp to 1000 bp with left, right, or double size selection
INSTRUCTION MANUAL Select-a-Size DNA Clean & Concentrator MagBead Kit Catalog No. D4084 & D4085 Highlights Tunable: Size selection can be tuned from 100 bp to 1000 bp with left, right, or double size selection
Procedure & Checklist bp Amplicon Library Preparation and Sequencing
 Procedure & Checklist - 250 bp Amplicon Library Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit. This procedure is optimized for SMRTbell
Procedure & Checklist - 250 bp Amplicon Library Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit. This procedure is optimized for SMRTbell
Procedure & Checklist bp Template Preparation and Sequencing
 Procedure & Checklist - 500 bp Template Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit. This procedure is optimized for SMRTbell template
Procedure & Checklist - 500 bp Template Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit. This procedure is optimized for SMRTbell template
VAHTS Stranded mrna-seq Library Prep Kit for Illumina
 Instruction Manual VAHTS Stranded mrna-seq Library Prep Kit for Illumina Vazyme Cat #NR602 Vazyme Biotech Co., Ltd Web: www.vazyme.com Tel: 400-600-9335 Sales: Sales@vazyme.com Support: Support@ vazyme.com
Instruction Manual VAHTS Stranded mrna-seq Library Prep Kit for Illumina Vazyme Cat #NR602 Vazyme Biotech Co., Ltd Web: www.vazyme.com Tel: 400-600-9335 Sales: Sales@vazyme.com Support: Support@ vazyme.com
Procedure & Checklist - 10 kb Template Preparation and Sequencing (with Low-Input DNA)
 Procedure & Checklist - 10 kb Template Preparation and Sequencing (with Low-Input DNA) Before You Begin To perform this procedure, you must have the PacBio : Template Prep Kit DNA/Polymerase Binding Kit
Procedure & Checklist - 10 kb Template Preparation and Sequencing (with Low-Input DNA) Before You Begin To perform this procedure, you must have the PacBio : Template Prep Kit DNA/Polymerase Binding Kit
Procedure & Checklist - 10 kb Template Preparation and Sequencing
 Procedure & Checklist - 10 kb Template Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit. This procedure can be used to prepare 10 kb libraries
Procedure & Checklist - 10 kb Template Preparation and Sequencing Before You Begin To perform this procedure, you must have the PacBio Template Prep Kit. This procedure can be used to prepare 10 kb libraries
10 kb to 20 kb Template Preparation and Sequencing with Low-Input DNA
 Please note: the shared protocols described herein may not have been validated by Pacific Biosciences and are provided as-is and without any warranty. Use of these protocols is offered to those customers
Please note: the shared protocols described herein may not have been validated by Pacific Biosciences and are provided as-is and without any warranty. Use of these protocols is offered to those customers
AGENCOURT ORAPURE Buccal Cell DNA Isolation Kit
 Buccal Cell DNA Isolation Kit Page 1 of 12 Please refer to http://www.agencourt.com/technical for updated protocols and refer to MSDS instructions when handling or shipping any chemical hazards. AGENCOURT
Buccal Cell DNA Isolation Kit Page 1 of 12 Please refer to http://www.agencourt.com/technical for updated protocols and refer to MSDS instructions when handling or shipping any chemical hazards. AGENCOURT
mi-mag mrna Isolation Kit
 mi-mag mrna Isolation Kit Cat. No [50 Reactions] This kit is for research purposes only. Not for use in diagnostic procedures. For in vitro use only. Introduction This kit contains enough materials for
mi-mag mrna Isolation Kit Cat. No [50 Reactions] This kit is for research purposes only. Not for use in diagnostic procedures. For in vitro use only. Introduction This kit contains enough materials for
Mag-Bind cfdna Kit. M preps M preps M Preps
 Mag-Bind cfdna Kit M3298-00 5 preps M3298-01 50 preps M3298-02 200 Preps March 2018 Mag-Bind cfdna Kit Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Preparing Reagents...4
Mag-Bind cfdna Kit M3298-00 5 preps M3298-01 50 preps M3298-02 200 Preps March 2018 Mag-Bind cfdna Kit Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Preparing Reagents...4
Procedure & Checklist - Greater Than 10 kb Template Preparation Using AMPure PB Beads
 Procedure & Checklist - Greater Than 10 kb Template Preparation Using AMPure PB Beads Before You Begin This procedure can be used to prepare greater than 10 kb libraries from 5 μg of sheared and concentrated
Procedure & Checklist - Greater Than 10 kb Template Preparation Using AMPure PB Beads Before You Begin This procedure can be used to prepare greater than 10 kb libraries from 5 μg of sheared and concentrated
Procedure & Checklist - cdna Capture Using SeqCap EZ Libraries
 Procedure & Checklist - cdna Capture Using SeqCap EZ Libraries Before You Begin This document describes the process for capturing cdna prepared with the SMARTer PCR cdna Synthesis Kit (Clontech) and pulled-down
Procedure & Checklist - cdna Capture Using SeqCap EZ Libraries Before You Begin This document describes the process for capturing cdna prepared with the SMARTer PCR cdna Synthesis Kit (Clontech) and pulled-down
NR601. VAHTS TM mrna-seq V2 Library Prep Kit for Illumina
 NR601 VAHTS TM mrna-seq V2 Library Prep Kit for Illumina v Vazyme Biotech Co., Ltd Website: www.vazyme.com Order: global@vazyme.com Support: support@vazyme.com Service: service@vazyme.com SYSTEMS www.vazyme.com
NR601 VAHTS TM mrna-seq V2 Library Prep Kit for Illumina v Vazyme Biotech Co., Ltd Website: www.vazyme.com Order: global@vazyme.com Support: support@vazyme.com Service: service@vazyme.com SYSTEMS www.vazyme.com
illumina TruSeq RNA Sample Prep. (LT) Protocol 1
 illumina TruSeq RNA Sample Prep. (LT) Protocol 1 Performed using the TruSeq RNA Sample Preparation Kit (A cat#fc-122-1001, B cat#fc122-1002) Purify and Fragment mrna NOTE: Use 3ug of Total RNA to initiate
illumina TruSeq RNA Sample Prep. (LT) Protocol 1 Performed using the TruSeq RNA Sample Preparation Kit (A cat#fc-122-1001, B cat#fc122-1002) Purify and Fragment mrna NOTE: Use 3ug of Total RNA to initiate
GS FLX/Junior Titanium Technology
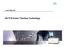 www.454.com GS FLX/Junior Titanium Technology GS FLX and GS Junior Process Steps Overview gdna 1. DNA Library Construction * 4h 2. empcr 3. Sequencing 8 h 10 h Data output DNA Library Preparation Prepare
www.454.com GS FLX/Junior Titanium Technology GS FLX and GS Junior Process Steps Overview gdna 1. DNA Library Construction * 4h 2. empcr 3. Sequencing 8 h 10 h Data output DNA Library Preparation Prepare
Procedure & Checklist Preparing SMRTbell Libraries using PacBio Barcoded Adapters for Multiplex SMRT Sequencing
 Procedure & Checklist Preparing SMRTbell Libraries using PacBio Barcoded Adapters for Multiplex SMRT Sequencing Before You Begin This document describes a procedure for multiplexing 5 Mb microbial genomes
Procedure & Checklist Preparing SMRTbell Libraries using PacBio Barcoded Adapters for Multiplex SMRT Sequencing Before You Begin This document describes a procedure for multiplexing 5 Mb microbial genomes
Additional reagents and materials that are not supplied
 sparq PureMag Beads Cat. No. 95196-005 Size: 5 ml Store at 2 C to 8 C 95196-060 60 ml 95196-450 450 ml Description sparq PureMag Beads uses reversible nucleic acid-binding properties of magnetic beads
sparq PureMag Beads Cat. No. 95196-005 Size: 5 ml Store at 2 C to 8 C 95196-060 60 ml 95196-450 450 ml Description sparq PureMag Beads uses reversible nucleic acid-binding properties of magnetic beads
Procedure & Checklist - Preparing SMRTbell Libraries using PacBio Barcoded Adapters for Multiplex SMRT Sequencing
 Procedure & Checklist - Preparing SMRTbell Libraries using PacBio Barcoded Adapters for Multiplex SMRT Sequencing Before You Begin This document describes methods for generating SMRTbell libraries using
Procedure & Checklist - Preparing SMRTbell Libraries using PacBio Barcoded Adapters for Multiplex SMRT Sequencing Before You Begin This document describes methods for generating SMRTbell libraries using
USER GUIDE For Illumina Platform
 USER GUIDE For Illumina Platform Copyright Nimagen B.V. P.O. Box 91 6500 AB Nijmegen The Netherlands Tel. +31 (0)24 820 0241 Fax. +31 (0)24 358 0259 info@nimagen.com VAT#: NL850011243B01 Rabobank Nijmegen:
USER GUIDE For Illumina Platform Copyright Nimagen B.V. P.O. Box 91 6500 AB Nijmegen The Netherlands Tel. +31 (0)24 820 0241 Fax. +31 (0)24 358 0259 info@nimagen.com VAT#: NL850011243B01 Rabobank Nijmegen:
chemagic mrna Direct Kit
 chemagic mrna Direct Kit for general purposes Kit for the direct isolation of mrna from animal and plant tissue and cells. Kit Components M-PVA OdT Magnetic Beads Suspension Buffer 1 Lysis Buffer 2 Wash
chemagic mrna Direct Kit for general purposes Kit for the direct isolation of mrna from animal and plant tissue and cells. Kit Components M-PVA OdT Magnetic Beads Suspension Buffer 1 Lysis Buffer 2 Wash
Procedure & Checklist - Iso-Seq Template Preparation for Sequel Systems
 Procedure & Checklist - Iso-Seq Template Preparation for Sequel Systems Before You Begin The long read lengths of the PacBio System are well-suited for characterizing full-length transcripts produced from
Procedure & Checklist - Iso-Seq Template Preparation for Sequel Systems Before You Begin The long read lengths of the PacBio System are well-suited for characterizing full-length transcripts produced from
Illumina TruSeq Stranded mrna (LT) Protocol 1
 Illumina TruSeq Stranded mrna (LT) Protocol 1 Performed using the TruSeq Stranded mrna Sample Preparation Kit (A cat#fc-122-2101, B cat#fc122-2102) Purify and Fragment mrna NOTE: Use 500ng of Total RNA
Illumina TruSeq Stranded mrna (LT) Protocol 1 Performed using the TruSeq Stranded mrna Sample Preparation Kit (A cat#fc-122-2101, B cat#fc122-2102) Purify and Fragment mrna NOTE: Use 500ng of Total RNA
30 Plex Human Luminex (Invitrogen Kit, Single Plate)
 30 Plex Human Luminex (Invitrogen Kit, Single Plate) 1. Defrost samples and bring to room temperature. 2. Bring Kit components to room temperature: Wash solution 20x. Assay Diluent. Incubation buffer.
30 Plex Human Luminex (Invitrogen Kit, Single Plate) 1. Defrost samples and bring to room temperature. 2. Bring Kit components to room temperature: Wash solution 20x. Assay Diluent. Incubation buffer.
pluribead KIT Cell Separation Protocol M-Bead
 pluribead KIT Cell Separation Protocol M-Bead pluriselect@hiss-dx.de 15 14 13 12 11 9 8 7 6 5 4 3 2 1 50 40 30 20 2 Contents Contents pluribead Kit Components & Additional Materials 2 Separation Protocol
pluribead KIT Cell Separation Protocol M-Bead pluriselect@hiss-dx.de 15 14 13 12 11 9 8 7 6 5 4 3 2 1 50 40 30 20 2 Contents Contents pluribead Kit Components & Additional Materials 2 Separation Protocol
MinION PROTOCOL. Adapted from Janneke Wit by Robyn Tanny May Company Kit/Item Catalog Number
 MinION PROTOCOL Adapted from Janneke Wit by Robyn Tanny May 2016 Company Kit/Item Catalog Number Fisher Eppendorf DNA/RNA LoBind Tubes 13-698-791 Fisher Covaris g-tube NC0380758 NEB NEBNext FFPE Repair
MinION PROTOCOL Adapted from Janneke Wit by Robyn Tanny May 2016 Company Kit/Item Catalog Number Fisher Eppendorf DNA/RNA LoBind Tubes 13-698-791 Fisher Covaris g-tube NC0380758 NEB NEBNext FFPE Repair
Microwell-Seq. High-throughput Single Cell RNA-Seq Kit. Protocol. Index
 Microwell-Seq High-throughput Single Cell RNA-Seq Kit Protocol Index 1 Introduction 2 Kit Reagent 3 Store 4 Application 5 Prepared materials 6 Note 7 Preparation 8 Workflow 1 1 Introduction Microwell-Seq
Microwell-Seq High-throughput Single Cell RNA-Seq Kit Protocol Index 1 Introduction 2 Kit Reagent 3 Store 4 Application 5 Prepared materials 6 Note 7 Preparation 8 Workflow 1 1 Introduction Microwell-Seq
JetSeq Clean. Product Manual
 JetSeq Clean Product Manual 2 Product Manual bioline.com/jetseq JetSeq Clean JetSeq Clean TABLE OF CONTENTS 1 Kit contents 04 2 Description 05 3 Equipment and reagents to be supplied by user 06 4 Storage
JetSeq Clean Product Manual 2 Product Manual bioline.com/jetseq JetSeq Clean JetSeq Clean TABLE OF CONTENTS 1 Kit contents 04 2 Description 05 3 Equipment and reagents to be supplied by user 06 4 Storage
Controller Training Kit User Guide
 Library Construction Chromium Controller Training Kit User Guide FOR USE WITH Chromium Controller & Training Reagents, Gel Bead & Chip Kits PN-120245 NOTICES Notices Manual Part Number CG00021 Rev A Legal
Library Construction Chromium Controller Training Kit User Guide FOR USE WITH Chromium Controller & Training Reagents, Gel Bead & Chip Kits PN-120245 NOTICES Notices Manual Part Number CG00021 Rev A Legal
BoTest Matrix E Botulinum Neurotoxin Detection Kit Protocol
 BoTest Matrix E Botulinum Neurotoxin Detection Kit Protocol 505 S. Rosa Road, Suite 105 Madison, WI 53719 1-608-441-8174 info@biosentinelpharma.com BioSentinel Part No: L1016, Release Date: May 29, 2014
BoTest Matrix E Botulinum Neurotoxin Detection Kit Protocol 505 S. Rosa Road, Suite 105 Madison, WI 53719 1-608-441-8174 info@biosentinelpharma.com BioSentinel Part No: L1016, Release Date: May 29, 2014
Procedure & Checklist - Preparing >15 kb Libraries Using SMRTbell Express Template Preparation Kit
 Procedure & Checklist - Preparing >15 kb Libraries Using SMRTbell Express Template Preparation Kit This document provides recommendations for preparing >15 kb size-selected SMRTbell libraries from 3-5
Procedure & Checklist - Preparing >15 kb Libraries Using SMRTbell Express Template Preparation Kit This document provides recommendations for preparing >15 kb size-selected SMRTbell libraries from 3-5
5. Carefully remove the printer from the lower boxed foam support and place it on a solid, level base where it will be used
 PROJET 1200 QUICKSTART GUIDE Before you get started you will need: Lint-free paper towels A pair of nitrile gloves Saftety glasses OPENING YOUR PROJET 1200 NOTE: Make sure you save all of your packaging
PROJET 1200 QUICKSTART GUIDE Before you get started you will need: Lint-free paper towels A pair of nitrile gloves Saftety glasses OPENING YOUR PROJET 1200 NOTE: Make sure you save all of your packaging
NGS clean-up and size selection
 NGS clean-up and size selection User manual NucleoMag NGS Clean-up and Size Select May 2014 / Rev. 01 NGS clean-up and size selection Table of contents 1 Components 4 1.1 Kit contents 4 1.2 Equipment and
NGS clean-up and size selection User manual NucleoMag NGS Clean-up and Size Select May 2014 / Rev. 01 NGS clean-up and size selection Table of contents 1 Components 4 1.1 Kit contents 4 1.2 Equipment and
TrueBlot Protein G Magnetic Beads PG ml. TrueBlot Protein G Magnetic Beads PG ml. Bead Mean Diameter 0.5 µm. Bead Concentration
 Rockland s TrueBlot Protein G Magnetic Beads are uniform, non-aggregating, super-paramagnetic beads coupled with a biomolecule, such as Protein G. These beads are specifically designed, tested and quality
Rockland s TrueBlot Protein G Magnetic Beads are uniform, non-aggregating, super-paramagnetic beads coupled with a biomolecule, such as Protein G. These beads are specifically designed, tested and quality
Procedure & Checklist cdna Capture Using IDT xgen Lockdown Probes
 Procedure & Checklist cdna Capture Using IDT xgen Lockdown Probes Before You Begin This document describes the process for capturing cdna prepared with the SMARTer PCR cdna Synthesis Kit (Clontech) and
Procedure & Checklist cdna Capture Using IDT xgen Lockdown Probes Before You Begin This document describes the process for capturing cdna prepared with the SMARTer PCR cdna Synthesis Kit (Clontech) and
We want to thank and acknowledge the authors for sharing this protocol and their contributions to the field.
 We adopted the protocol described in the Extended Experimental Procedures section I.a.1 of the 2014 Cell paper by Rao and Huntley et. al: A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles
We adopted the protocol described in the Extended Experimental Procedures section I.a.1 of the 2014 Cell paper by Rao and Huntley et. al: A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles
HP 564 and 920 InkJet Cartridges Refill Instructions (Professional Version)
 HP 564 and 920 InkJet Cartridges Refill Instructions (Professional Version) For the following cartridges: 934, 934XL, 935, and 935XL Series 5869 Terminal Ave. I Colorado Springs, CO 80915 PH: 719-578-0506
HP 564 and 920 InkJet Cartridges Refill Instructions (Professional Version) For the following cartridges: 934, 934XL, 935, and 935XL Series 5869 Terminal Ave. I Colorado Springs, CO 80915 PH: 719-578-0506
DNA Size Selection Magnetic Beads
 DNA Size Selection Magnetic Beads Catalog #: 801-117 User Manual Last revised July 30 th, 2018 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100 Norcross,
DNA Size Selection Magnetic Beads Catalog #: 801-117 User Manual Last revised July 30 th, 2018 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100 Norcross,
ChIP Protocol for fresh or frozen cross linked cells
 Prior to starting your ChIPs and Shearing Turn on sonifiers and cooling system allow system to reach -2 C before shearing Cool bench top centrifuge to 4 C Prepare all of your buffers with protease inhibitors
Prior to starting your ChIPs and Shearing Turn on sonifiers and cooling system allow system to reach -2 C before shearing Cool bench top centrifuge to 4 C Prepare all of your buffers with protease inhibitors
RayBio anti-mouse IgG Magnetic Beads
 RayBio anti-mouse IgG Magnetic Beads Catalog #: 801-103 User Manual Last revised January 4 th, 2017 Caution: Extraordinarily useful information enclosed ISO 1348 Certified 3607 Parkway Lane, Suite 100
RayBio anti-mouse IgG Magnetic Beads Catalog #: 801-103 User Manual Last revised January 4 th, 2017 Caution: Extraordinarily useful information enclosed ISO 1348 Certified 3607 Parkway Lane, Suite 100
Target Sequence Capture Using Roche NimbleGen SeqCap EZ Library
 Please note: the shared protocols described herein may not have been validated by Pacific Biosciences and are provided as-is and without any warranty. Use of these protocols is offered to those customers
Please note: the shared protocols described herein may not have been validated by Pacific Biosciences and are provided as-is and without any warranty. Use of these protocols is offered to those customers
Corrie Moreau Page 1 7/23/10
 Corrie Moreau Page 1 7/23/10 DNA Extractions using Qiagen DNeasy Kits with Extraction Beads Corrie Moreau Field Museum (September 2009) These are the instructions I use for DNA extractions of individual
Corrie Moreau Page 1 7/23/10 DNA Extractions using Qiagen DNeasy Kits with Extraction Beads Corrie Moreau Field Museum (September 2009) These are the instructions I use for DNA extractions of individual
Formaldehyde Cross-linking of Chromatin from Drosophila
 2 Formaldehyde Cross-linking of Chromatin from Drosophila Protocol from modencode IGSB University of Chicago originally written by Alex Crofts and Sasha Ostapenko and updated by Matt Kirkey. 1. Set centrifuge
2 Formaldehyde Cross-linking of Chromatin from Drosophila Protocol from modencode IGSB University of Chicago originally written by Alex Crofts and Sasha Ostapenko and updated by Matt Kirkey. 1. Set centrifuge
Required Materials. Page 1 PN Version 06 (February 2018)
 Procedure & Checklist - Preparing >30 kb SMRTbell Libraries Using Megaruptor Shearing and BluePippin Size-Selection for PacBio RS II and Sequel Systems This document provides recommendations for preparing
Procedure & Checklist - Preparing >30 kb SMRTbell Libraries Using Megaruptor Shearing and BluePippin Size-Selection for PacBio RS II and Sequel Systems This document provides recommendations for preparing
RayBio mrna Magnetic Beads Kit
 RayBio mrna Magnetic Beads Kit Catalog #: 801-116 User Manual Last revised March 9 th, 2017 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100 Norcross,
RayBio mrna Magnetic Beads Kit Catalog #: 801-116 User Manual Last revised March 9 th, 2017 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100 Norcross,
Directions for running IC samples on the Shimadzu TOC analyzer. (5/18/2015 DER)
 Directions for running IC samples on the Shimadzu TOC analyzer. (5/18/2015 DER) Turn on the TOC analyzer by pressing the on switch located in the lower left corner of the panel on the front of the instrument.
Directions for running IC samples on the Shimadzu TOC analyzer. (5/18/2015 DER) Turn on the TOC analyzer by pressing the on switch located in the lower left corner of the panel on the front of the instrument.
Procedure & Checklist Multiplex Genomic DNA Target Capture Using IDT xgen Lockdown Probes
 Procedure & Checklist Multiplex Genomic DNA Target Capture Using IDT xgen Lockdown Probes Before You Begin This procedure describes capture and enrichment of regions of interest by using IDT xgen Lockdown
Procedure & Checklist Multiplex Genomic DNA Target Capture Using IDT xgen Lockdown Probes Before You Begin This procedure describes capture and enrichment of regions of interest by using IDT xgen Lockdown
AdnaTest EMT-1/StemCellSelect
 AdnaTest EMT-1/StemCellSelect Enrichment of tumor cells from blood for gene expression analysis For research use only Manual T-1-533 Contents Order Information... 3 Purpose... 3 Abbreviations and Symbols...
AdnaTest EMT-1/StemCellSelect Enrichment of tumor cells from blood for gene expression analysis For research use only Manual T-1-533 Contents Order Information... 3 Purpose... 3 Abbreviations and Symbols...
MEDIP-SEQUENCING PROTOCOL
 MEDIP-SEQUENCING PROTOCOL MAGMEDIP KIT Cat. No. C02010020 Table 1 The GenDNA module provides you with an excess of buffer for the preparation of DNA. Sufficient buffer is given for the preparation of several
MEDIP-SEQUENCING PROTOCOL MAGMEDIP KIT Cat. No. C02010020 Table 1 The GenDNA module provides you with an excess of buffer for the preparation of DNA. Sufficient buffer is given for the preparation of several
EASY-IN POOL STEP SYSTEM NE132
 EASY-IN POOL STEP SYSTEM NE132 This instruction manual features multiple guides for the step unit components. 7939 EASY POOL STEP (NE113) FOR USE WITH: EASY-IN POOL STEP (NE126) 6492 PARTS & HARDWARE FOR
EASY-IN POOL STEP SYSTEM NE132 This instruction manual features multiple guides for the step unit components. 7939 EASY POOL STEP (NE113) FOR USE WITH: EASY-IN POOL STEP (NE126) 6492 PARTS & HARDWARE FOR
Alon s SCN ChIP Protocol
 Prior to starting your ChIPs and Shearing 1. Turn on sonifiers and cooling system allow system to reach -1 C before shearing 2. Cool bench top centrifuge to 4 C 3. Prepare all of your buffers with protease
Prior to starting your ChIPs and Shearing 1. Turn on sonifiers and cooling system allow system to reach -1 C before shearing 2. Cool bench top centrifuge to 4 C 3. Prepare all of your buffers with protease
Procedure & Checklist Preparing Multiplexed Microbial SMRTbell Libraries for the PacBio Sequel System
 Procedure & Checklist Preparing Multiplexed Microbial SMRTbell Libraries for the PacBio Sequel System Before You Begin This procedure is for preparing multiplexed SMRTbell libraries for sequencing on the
Procedure & Checklist Preparing Multiplexed Microbial SMRTbell Libraries for the PacBio Sequel System Before You Begin This procedure is for preparing multiplexed SMRTbell libraries for sequencing on the
just below the screen. Data collection will begin, and a graph will show your data being plotted in real time.
 To Collect Additional Data To start a second data collection run, tap the file should now see Run 2 displayed with a blank graph. cabinet in the upper right corner. You just below the screen. Data collection
To Collect Additional Data To start a second data collection run, tap the file should now see Run 2 displayed with a blank graph. cabinet in the upper right corner. You just below the screen. Data collection
AdnaTest OvarianCancer-2 Select
 AdnaTest OvarianCancer-2 Select Enrichment of tumor cells from blood of ovarian cancer patients for gene expression analysis For research use only Manual T-1-538 Contents Order Information... 3 Purpose...
AdnaTest OvarianCancer-2 Select Enrichment of tumor cells from blood of ovarian cancer patients for gene expression analysis For research use only Manual T-1-538 Contents Order Information... 3 Purpose...
JetSeq DNA Library Preparation Kit. Product Manual
 JetSeq DNA Library Preparation Kit Product Manual 2 JetSeq DNA Library Preparation Kit JetSeq DNA Library Preparation Kit TABLE OF CONTENTS 1 Kit contents 04 2 Description 05 3 Storage 06 4 Safety information
JetSeq DNA Library Preparation Kit Product Manual 2 JetSeq DNA Library Preparation Kit JetSeq DNA Library Preparation Kit TABLE OF CONTENTS 1 Kit contents 04 2 Description 05 3 Storage 06 4 Safety information
BIOL110L-Cell Biology Lab Spring Quarter 2012 Module 3-4 Wednesday May 30, 2012
 BIOL110L-Cell Biology Lab Spring Quarter 2012 Module 3-4 Wednesday May 30, 2012 PART I: Isolation of over-expressed GFP-Karyopherins from yeast extracts by affinity capture on nucleoporins Summary: The
BIOL110L-Cell Biology Lab Spring Quarter 2012 Module 3-4 Wednesday May 30, 2012 PART I: Isolation of over-expressed GFP-Karyopherins from yeast extracts by affinity capture on nucleoporins Summary: The
Quick Reference Guide
 Quick Reference Guide Table of Contents Defoamer - Composite Blocks...1 Zirlux FC2...1 Milling a Restoration...2 Tool Gauges and Warnings...3 Automatic Tool Changer...4 Maintenance Reminders...5 Clean/Refill
Quick Reference Guide Table of Contents Defoamer - Composite Blocks...1 Zirlux FC2...1 Milling a Restoration...2 Tool Gauges and Warnings...3 Automatic Tool Changer...4 Maintenance Reminders...5 Clean/Refill
LOABeads MagSep 15/50 LOABeads MagSep 500
 LOABeads MagSep 15/50 LOABeads MagSep 500 Product Manual Lab on a Bead AB Edition 2016-05-09 Copyright 2015-2016 Lab on a Bead AB Table of Contents 1. 2. 3. 4. 5. 6. Safety instructions...3 General handling
LOABeads MagSep 15/50 LOABeads MagSep 500 Product Manual Lab on a Bead AB Edition 2016-05-09 Copyright 2015-2016 Lab on a Bead AB Table of Contents 1. 2. 3. 4. 5. 6. Safety instructions...3 General handling
NEBNext DNA Library Prep Master Mix Set for Illumina
 LIBRARY PREPARATION NEBNext DNA Library Prep Master Mix Set for Illumina Instruction Manual NEB #E6040S/L 12/60 reactions Version 8.0 9/18 be INSPIRED drive DISCOVERY stay GENUINE This product is intended
LIBRARY PREPARATION NEBNext DNA Library Prep Master Mix Set for Illumina Instruction Manual NEB #E6040S/L 12/60 reactions Version 8.0 9/18 be INSPIRED drive DISCOVERY stay GENUINE This product is intended
mag maxi kit Intended use of the mag maxi kits
 mag maxi kit For in vitro diagnostic use 40403 40430 10 288 May 2014 LGC Genomics GmbH Ostendstr. 25 TGS Haus 8 12459 Berlin Germany Tel: +49 (0)30 5304 2200 Fax: +49 (0)30 5304 2201 Intended use of the
mag maxi kit For in vitro diagnostic use 40403 40430 10 288 May 2014 LGC Genomics GmbH Ostendstr. 25 TGS Haus 8 12459 Berlin Germany Tel: +49 (0)30 5304 2200 Fax: +49 (0)30 5304 2201 Intended use of the
EASY POOL STEP (NE113)
 EASY POOL STEP (NE113) FOR USE WITH: EASY POOL STEP (NE113) (1 CARTON) EASY POOL STEP WITH OUTSIDE LADDER (NE126) EASY POOL STEP ENTRY SYSTEM (NE138) (With Gate) (4 CARTONS) Above are the options available
EASY POOL STEP (NE113) FOR USE WITH: EASY POOL STEP (NE113) (1 CARTON) EASY POOL STEP WITH OUTSIDE LADDER (NE126) EASY POOL STEP ENTRY SYSTEM (NE138) (With Gate) (4 CARTONS) Above are the options available
Procedure & Checklist - Iso-Seq Template Preparation for Sequel Systems
 Procedure & Checklist - Iso-Seq Template Preparation for Sequel Systems Before You Begin The Sequel System generates long reads that are well-suited for characterizing full-length transcripts produced
Procedure & Checklist - Iso-Seq Template Preparation for Sequel Systems Before You Begin The Sequel System generates long reads that are well-suited for characterizing full-length transcripts produced
ChargeSwitch gdna Rendered Meat Purification Kit
 USER GUIDE ChargeSwitch gdna Rendered Meat Purification Kit Purification of genomic DNA (gdna) from cattle feed, meal, and heparin products Catalog Number CS400-100 Publication Number MAN0000574 Revision
USER GUIDE ChargeSwitch gdna Rendered Meat Purification Kit Purification of genomic DNA (gdna) from cattle feed, meal, and heparin products Catalog Number CS400-100 Publication Number MAN0000574 Revision
PARP1-Trap_A for Immunoprecipitation of PARP1- Fusion Proteins from cell extract
 PARP1-Trap_A for Immunoprecipitation of PARP1- Fusion Proteins from cell extract Only for research applications, not for diagnostic or therapeutic use. Introduction Specificity Poly(ADP-ribose) polymerase
PARP1-Trap_A for Immunoprecipitation of PARP1- Fusion Proteins from cell extract Only for research applications, not for diagnostic or therapeutic use. Introduction Specificity Poly(ADP-ribose) polymerase
SOP-P094. BioMek 2000 Compound Protocol Cyan/Hypercyt + Analysis
 SOP-P094 BioMek 2000 Compound Protocol Cyan/Hypercyt + Analysis Objective: To test serial dilutions of certain compounds Protocol for half plate runs only. One plate of Redox dye and the other plate -
SOP-P094 BioMek 2000 Compound Protocol Cyan/Hypercyt + Analysis Objective: To test serial dilutions of certain compounds Protocol for half plate runs only. One plate of Redox dye and the other plate -
HP Color LaserJet CP3525 Series Manage and maintain
 Load paper and print media Load Tray 1 1 Open Tray 1. CAUTION: To avoid jams, never add or remove paper from Tray 1 during printing. 2 Fold out the tray extension to support the paper and set the side
Load paper and print media Load Tray 1 1 Open Tray 1. CAUTION: To avoid jams, never add or remove paper from Tray 1 during printing. 2 Fold out the tray extension to support the paper and set the side
TITLE: GUIDELINES FOR SAMPLE COLLECTION U + B
 TITLE: GUIDELINES FOR SAMPLE COLLECTION U + B 1 THE ARFL SAMPLE AND BARCODED SECURITY DOCUMENTATION SYSTEM 1.1 Range of Sampling Kits The small Sampling Kit contains one urine sample pack and one blood
TITLE: GUIDELINES FOR SAMPLE COLLECTION U + B 1 THE ARFL SAMPLE AND BARCODED SECURITY DOCUMENTATION SYSTEM 1.1 Range of Sampling Kits The small Sampling Kit contains one urine sample pack and one blood
Replacing the print cartridges
 http://www.hp.com/support/lj9500 1 2 3 Replacing the print cartridges The printer uses four color print cartridges: yellow, magenta, cyan, and black. Follow this procedure to install the print cartridges.
http://www.hp.com/support/lj9500 1 2 3 Replacing the print cartridges The printer uses four color print cartridges: yellow, magenta, cyan, and black. Follow this procedure to install the print cartridges.
EASY POOL STEP (NE113)
 EASY POOL STEP (NE113) FOR USE WITH: EASY POOL STEP (NE113) (1 CARTON) EASY POOL STEP WITH OUTSIDE LADDER (NE126) EASY POOL STEP ENTRY SYSTEM (NE138) (With Gate) (4 CARTONS) Above are the options available
EASY POOL STEP (NE113) FOR USE WITH: EASY POOL STEP (NE113) (1 CARTON) EASY POOL STEP WITH OUTSIDE LADDER (NE126) EASY POOL STEP ENTRY SYSTEM (NE138) (With Gate) (4 CARTONS) Above are the options available
FastTrack MAG mrna Isolation Kits
 USER GUIDE FastTrack MAG mrna Isolation Kits For isolating high-quality mrna from total RNA, cells, and tissue Catalog Numbers K1580-01 and K1580-02 Document Part Number 25-0754 Publication Number MAN0000475
USER GUIDE FastTrack MAG mrna Isolation Kits For isolating high-quality mrna from total RNA, cells, and tissue Catalog Numbers K1580-01 and K1580-02 Document Part Number 25-0754 Publication Number MAN0000475
Installation Instructions 8115F 8115SF
 TM Installation Instructions 85F 85SF Single Control Centerset Lavatory Faucet with Speed Connect Drain Congratulations on purchasing your American Standard faucet with the Speed Connect Drain, a feature
TM Installation Instructions 85F 85SF Single Control Centerset Lavatory Faucet with Speed Connect Drain Congratulations on purchasing your American Standard faucet with the Speed Connect Drain, a feature
xgen hybridization capture of DNA libraries
 xgen hybridization capture of DNA libraries For NGS target enrichment Uses IDT s: xgen Hybridization and Wash Kit xgen Universal Blockers TS Mix, 10 bp TS Mix, or NXT Mix xgen Lockdown Panels and Probes
xgen hybridization capture of DNA libraries For NGS target enrichment Uses IDT s: xgen Hybridization and Wash Kit xgen Universal Blockers TS Mix, 10 bp TS Mix, or NXT Mix xgen Lockdown Panels and Probes
NxSeq UltraLow DNA Library Kit, 12 Reactions
 NxSeq UltraLow DNA Library Kit, 12 Reactions Illumina-compatible FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695
NxSeq UltraLow DNA Library Kit, 12 Reactions Illumina-compatible FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695
Project: RADseqReady Plate # Library # Name: Date: Section 1: DNA Standardization
 BestRAD Library Preparation Based on protocol of Ali et al. 2015 (10.1534/genetics.115.183665) Adapted by Linda Rutledge many times but this version was done on August 16, 2016 Section 1: DNA Standardization
BestRAD Library Preparation Based on protocol of Ali et al. 2015 (10.1534/genetics.115.183665) Adapted by Linda Rutledge many times but this version was done on August 16, 2016 Section 1: DNA Standardization
Enriching Beads Oligo (dt) Magnetic Beads for mrna Purification
 Enriching Beads Oligo (dt) Magnetic Beads for mrna Purification Isolate the mrna transcriptome in 15 minutes User Guidance Enriching Biotechnology Rev. 1.0 October 25th. 2018 Why choose Enriching Beads
Enriching Beads Oligo (dt) Magnetic Beads for mrna Purification Isolate the mrna transcriptome in 15 minutes User Guidance Enriching Biotechnology Rev. 1.0 October 25th. 2018 Why choose Enriching Beads
Applied Biosystems SOLiD 4 System Templated Bead Preparation Guide
 Applied Biosystems SOLiD 4 System Templated Bead Preparation Guide March 2010 Library Preparation Templated Bead Preparation Instrument Operation For Research Use Only. Not intended for any animal or human
Applied Biosystems SOLiD 4 System Templated Bead Preparation Guide March 2010 Library Preparation Templated Bead Preparation Instrument Operation For Research Use Only. Not intended for any animal or human
EPIGENTEK. EpiQuik Circulating Cell-Free DNA Isolation Kit. Base Catalog # P-1064 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Circulating Cell-Free DNA Isolation Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE Uses: The EpiQuik Circulating Cell-Free DNA Isolation Kit utilizes magnetic beads based sizefractionation
EpiQuik Circulating Cell-Free DNA Isolation Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE Uses: The EpiQuik Circulating Cell-Free DNA Isolation Kit utilizes magnetic beads based sizefractionation
Care and Use of the VP 407AM-N 96 Pin Magnetic Bead Extractor
 TECHNICAL NOTE 310 Care and Use of the VP 407AM-N 96 Pin Magnetic Bead Extractor Cover Plate VP 407AM-N-PCR (purchase separately) Magnetic Bead Extractor VP 407AM-N Loading Frame For Cover Plate (included)
TECHNICAL NOTE 310 Care and Use of the VP 407AM-N 96 Pin Magnetic Bead Extractor Cover Plate VP 407AM-N-PCR (purchase separately) Magnetic Bead Extractor VP 407AM-N Loading Frame For Cover Plate (included)
Stranded mrna-seq Lib Prep Kit for Illumina
 Stranded mrna-seq Lib Prep Kit for Illumina RK20301 (10ng-1ug Input Total RNA) (Illumina Compatible) C U G A www.abclonal.com version: N12G13v1.0 Contents 1.Introduction 01 2.Components 02 3.Additional
Stranded mrna-seq Lib Prep Kit for Illumina RK20301 (10ng-1ug Input Total RNA) (Illumina Compatible) C U G A www.abclonal.com version: N12G13v1.0 Contents 1.Introduction 01 2.Components 02 3.Additional
Basic Users Manual for Tecnai-F20 TEM
 Basic Users Manual for Tecnai-F20 TEM NB: This document contains my personal notes on the operating procedure of the Tecnai F20 and may be used as a rough guide for those new to the microscope. It may
Basic Users Manual for Tecnai-F20 TEM NB: This document contains my personal notes on the operating procedure of the Tecnai F20 and may be used as a rough guide for those new to the microscope. It may
ChargeSwitch gdna Blood Kits
 Instruction Manual ChargeSwitch gdna Blood Kits For purification of genomic DNA from small volumes of human blood Catalog nos. CS11000, CS11010, and CS11010-10 Version A 6 January 2005 25-0814 ii Table
Instruction Manual ChargeSwitch gdna Blood Kits For purification of genomic DNA from small volumes of human blood Catalog nos. CS11000, CS11010, and CS11010-10 Version A 6 January 2005 25-0814 ii Table
CABINETRY Assembly Instructions
 www.hdicabinetry.com Assembly Instructions TABLE OF CONTENTS Category Page(s) Section 1: Framed Series Base Cabinet Instructions Wall Cabinet Instructions Easy Reach Cabinet Instructions 1.01-1.04 1.05-1.06
www.hdicabinetry.com Assembly Instructions TABLE OF CONTENTS Category Page(s) Section 1: Framed Series Base Cabinet Instructions Wall Cabinet Instructions Easy Reach Cabinet Instructions 1.01-1.04 1.05-1.06
CNC Using the FlexiCam CNC and HMI Software. Guldbergsgade 29N, P0 E: T:
 CNC Using the FlexiCam CNC and HMI Software Guldbergsgade 29N, P0 E: makerlab@kea.dk T: +46 46 03 90 This grey box is the NC controller. Let s start by turning the red switch to the ON position, then press
CNC Using the FlexiCam CNC and HMI Software Guldbergsgade 29N, P0 E: makerlab@kea.dk T: +46 46 03 90 This grey box is the NC controller. Let s start by turning the red switch to the ON position, then press
Using Your Chip Priming Station
 s1 Using Your Chip Priming Station The Chip Priming Station, part number 5065-4401, is for use with the Agilent 2100 Bioanalyzer LabChip Kits. Refer to Figure 1 and 2 for a picture of the Chip Priming
s1 Using Your Chip Priming Station The Chip Priming Station, part number 5065-4401, is for use with the Agilent 2100 Bioanalyzer LabChip Kits. Refer to Figure 1 and 2 for a picture of the Chip Priming
CleanPlex UMI NGS Panel
 CleanPlex UMI NGS Panel User Guide This user guide is for the following products: CleanPlex UMI Lung Cancer Panel CleanPlex UMI Custom NGS Panel Get the latest user guide at: www.paragongenomics.com/product_documents/
CleanPlex UMI NGS Panel User Guide This user guide is for the following products: CleanPlex UMI Lung Cancer Panel CleanPlex UMI Custom NGS Panel Get the latest user guide at: www.paragongenomics.com/product_documents/
General Help. Last revised: Winter When I try to print something on the computer, it appears to work, but nothing comes out of the printer.
 General Help Last revised: Winter 2015 Problem Solution When I try to print something on the computer, it appears to work, but nothing comes out of the printer. See the next item. When I try to print something
General Help Last revised: Winter 2015 Problem Solution When I try to print something on the computer, it appears to work, but nothing comes out of the printer. See the next item. When I try to print something
LOABeads AffiAmino. Product Manual. Lab on a Bead AB. Revision date Copyright Lab on a Bead AB All rights reserved
 LOABeads AffiAmino Product Manual Lab on a Bead AB Revision date 2016-11-23 Copyright 2015-2016 Lab on a Bead AB All rights reserved Table of Contents 1. General information...3 2. Product data...4 3.
LOABeads AffiAmino Product Manual Lab on a Bead AB Revision date 2016-11-23 Copyright 2015-2016 Lab on a Bead AB All rights reserved Table of Contents 1. General information...3 2. Product data...4 3.
User s Guide. Slide Feeder for Flextight 949 and X5 Scanners English
 User s Guide Slide Feeder for Flextight 949 and X5 Scanners English Table of contents Introduction 3 I m p o r t a n t Wa r n i n g s a n d R e s t r i c t i o n s 3 Sy s t e m R e q u i r e m e n t s
User s Guide Slide Feeder for Flextight 949 and X5 Scanners English Table of contents Introduction 3 I m p o r t a n t Wa r n i n g s a n d R e s t r i c t i o n s 3 Sy s t e m R e q u i r e m e n t s
EPSON Stylus C80. Ink Cartridges. User Replaceable Parts. Media. 1/02 EPSON Stylus C80-1. Paper support. Left edge guide
 Printer Parts Accessories Left edge guide Paper support Ink Cartridges Cartridge Part number Printer cover Right edge guide Black Cyan Magenta Yellow T032120 T032220 T032320 T032420 User Replaceable Parts
Printer Parts Accessories Left edge guide Paper support Ink Cartridges Cartridge Part number Printer cover Right edge guide Black Cyan Magenta Yellow T032120 T032220 T032320 T032420 User Replaceable Parts
ISOFECAL for Beads Beating Manual (First edition)
 Fecal DNA Extraction Kit ISOFECAL for Beads Beating Manual (First edition) Code No. 315-06281 NIPPON GENE CO., LTD. Table of contents I Product description 1 II Contents of kit 1 III Storage 2 IV Precautions
Fecal DNA Extraction Kit ISOFECAL for Beads Beating Manual (First edition) Code No. 315-06281 NIPPON GENE CO., LTD. Table of contents I Product description 1 II Contents of kit 1 III Storage 2 IV Precautions
